DriverDBv4: A database for human cancer driver gene research
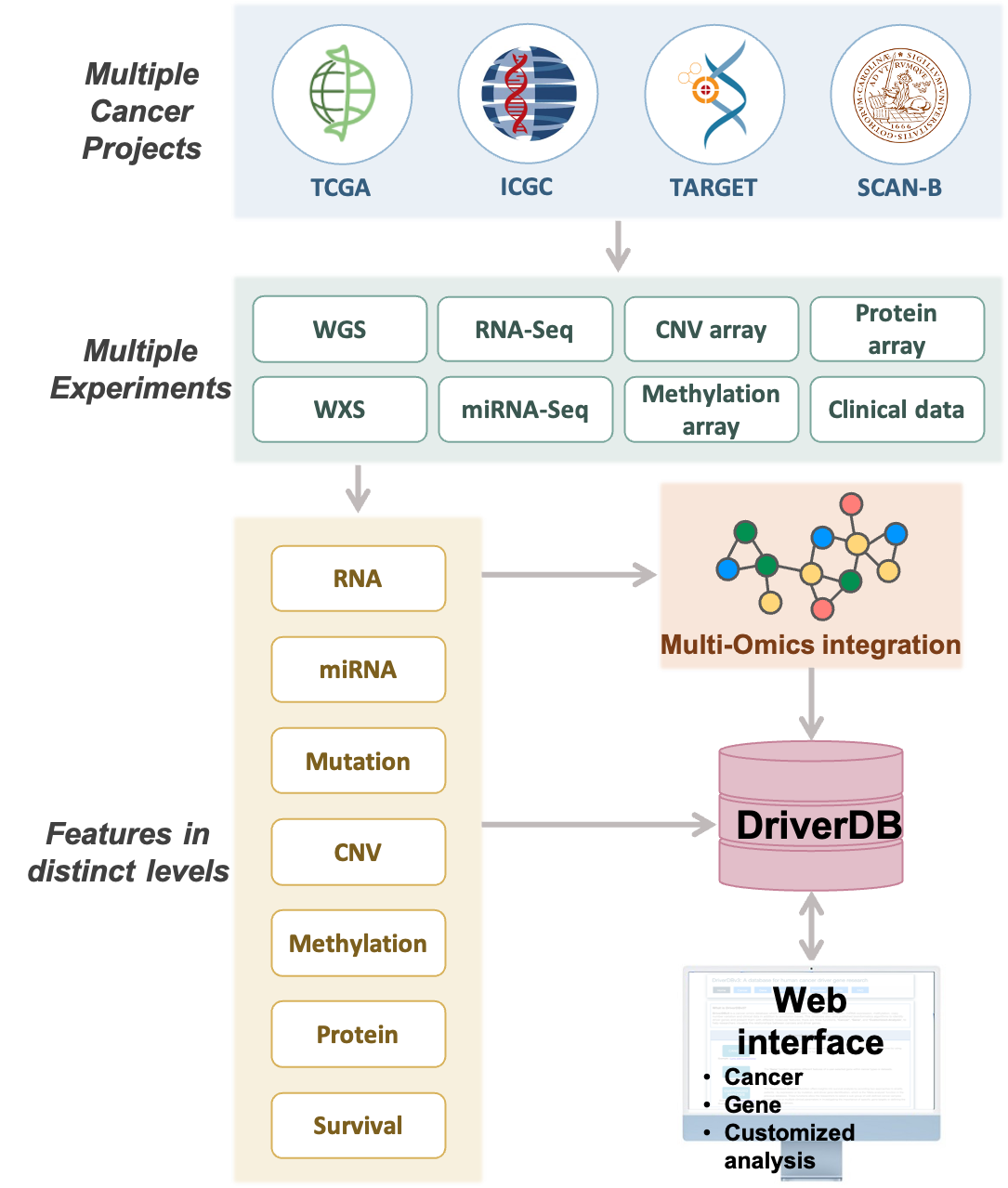
What is DriverDB?
DriverDB is an integrative cancer omics database that combines somatic mutation, RNA expression, miRNA expression, protein expression, methylation, copy number variation, and clinical data with curated annotations and published bioinformatics algorithms for driver gene and driver event identification. Featured in the 2014, 2016, and 2020 Nucleic Acids Research Database Issues, DriverDB applies state-of-the-art computational methods to characterize cancer drivers across molecular layers.
DriverDB provides three major analytical modules:- Cancer – Summarizes driver gene predictions for a selected cancer type across multiple omics layers using published driver identification tools.
- Gene – Visualizes multi-omics features of a user-selected gene, including differential expression, mutation, CNV, methylation, survival, miRNA regulation, protein expression, and integrated multi-omics evidence.
- Customized Analysis – Allows users to perform subgroup comparisons, survival analyses, multi-omics driver exploration, prognostic signature construction, and multivariate Cox modeling based on user-defined clinical or molecular criteria.
1. Cancer
1.1 Cancer Module Overview
The Cancer module summarizes driver gene and driver event predictions for a user-selected cancer type by integrating multi-omics data—including somatic mutations, RNA expression, miRNA expression, protein expression, copy number variation (CNV), methylation, and clinical information—through published bioinformatics algorithms and curated annotation sources. This module provides a cancer-centric overview of dysregulated molecular features and highlights candidate driver genes, their regulatory mechanisms, and their functional significance across molecular layers.1.2 Dataset Selection: Browse by Cancer Type
DriverDBv4 provides analysis across 70 cancer datasets, including 33 TCGA cancer types and additional datasets from resources such as CPTAC and ICGC. Use the selection panel to choose the dataset you want to explore.
A. Tissue Type (Optional)
Filter available datasets by tissue origin to quickly locate cancers related to a specific anatomical site.For example, selecting Lung narrows the list to datasets such as:
- Lung Adenocarcinoma (TCGA-US)
- Lung Squamous Cell Carcinoma (TCGA-US)
- Lung Cancer – KR (ICGC-KR)
B. Related Dataset
Select the specific cancer dataset you wish to analyze. Each dataset label includes its data source (e.g., TCGA-US, ICGC-KR), allowing users to choose cohorts most relevant to their research.
C. Submit
After making your selections, click Submit to load driver gene summaries and molecular features for the chosen cancer type. All downstream tabs, including Mutation, CNV, Methylation, Survival, miRNA, and Multi-Omics, will display results based on the selected dataset.

1.3 Overview of Result Tabs
The Cancer module contains several results tabs, each summarizing driver evidence derived from a different omics layer:- Summary – Integrates dysfunction and dysregulation evidence to highlight candidate driver genes and miRNA drivers for the selected cancer type.
- Mutation – Identifies mutation-based driver genes using multiple mutation detection tools.
- CNV – Visualizes driver genes with significant copy-number gain or loss, including CNV–expression relationships.
- Methylation – Highlights hyper- and hypomethylation driver genes and locus-enrichment distributions.
- Survival – Presents survival-relevant drivers and synergistic gene-pair interactions.
- miRNA – Shows regulatory interactions between differentially expressed genes and miRNAs.
- Multi-Omics – Integrates multiple omics layers to identify cross-omics driver genes and functional patterns.
1.4 Cancer Summary
1.4.1 Overview
The Cancer Summary tab provides an integrated overview of potential driver genes and miRNA drivers for the selected cancer type. It aggregates multi-omics driver evidence—including mutation, CNV, methylation, expression, miRNA regulation, and survival relevance—and connects them through known biological networks such as protein–protein interactions (PPIs), gene–miRNA interactions, and synergistic survival associations.
This section contains two main components:- Summary Network
- Driver Summary Table
Together, these views help users quickly identify influential driver genes, their regulatory relationships, and cross-omics support.
1.4.2 Summary Network
Purpose
The Summary Network visualizes the relationships between driver genes and miRNA drivers in the selected cancer type.
The network integrates:- Cancer Gene Census (CGC) annotations
- Network of Cancer Genes (NCG6.0) annotations
- Protein-protein interactions (PPIs) from STRING
- Synergistic effects, where the combined hazard ratio (HR) of two genes is >1.5× that of either individual gene
- miRNA–gene interactions from miRTarBase
This network highlights how multi-omics driver events relate to each other within a functional or regulatory context.
Nodes
Driver Gene Nodes- Displayed as colored circles
- Color indicates the omics type(s) contributing to driver status (see gene node legend)
- Red star inside node: gene identified by multi-omics driver tools

miRNA Driver Nodes
- Displayed as yellow nodes
- Represent miRNAs identified as regulatory or dysregulated in this cancer type
Edges
Each edge indicates a known or predicted biological relationship:
Unconnected nodes are removed to reduce visual clutter and highlight biologically relevant clusters.
Interaction Guide
The Summary Network is fully interactive:
Selecting and Highlighting- Click a node to highlight its connected genes/miRNAs and relationships
- Click blank space to return to the full network view
- Use the dropdown to jump directly to a specific gene of interest
- Gene Source: limit nodes to CGC genes, NCG genes, or all genes
- Node (Driver) Type: show only mutation, CNV, methylation, or miRNA-based drivers
- Interaction Type: show only PPIs, only synergistic effects, or both

1.4.3 Driver Summary Table
The Driver Summary Table lists all candidate driver genes for the selected cancer type, along with supporting evidence from each omics layer and known cancer gene resources.This table complements the network by providing a structured, quantitative summary of driver support.
1.5 Cancer Mutation
1.5.1 Overview
The Cancer Mutation section identifies and visualizes mutation-based driver genes in the selected cancer type. Driver genes are detected using multiple published mutation driver–prediction tools, and the degree of tool consensus provides a measure of confidence.
This section contains two components:- Mutation Driver Summary by Tools
- Mutation Profiles of Top 30 Driver Genes
Together, these views help users understand how many tools support each driver gene and how mutations are distributed across the patient cohort.
1.5.2 Mutation Driver Summary by Tools
Purpose
This panel summarizes how many genes are identified by varying numbers of mutation driver–detection tools.
Stronger consensus across tools indicates stronger evidence supporting a gene’s driver role.
Components
Distribution of Mutation Driver Genes by Tool Support (Left Plot)- Displays a bar plot showing the number of genes supported by 1, 2, 3… up to all mutation tools.
- Each bar represents how many driver genes were identified by that number of tools.
- Higher bars at larger tool counts indicate stronger multi-tool agreement.
Mutation Summary Table (Right Table)
- Located to the right of the plot.
- Lists the tool support count for each mutation driver gene.
- The plot on the left is derived from this table.

1.5.3 Mutation Profiles of Top 30 Driver Genes
Purpose
This section visualizes mutation patterns for the top 30 mutation driver genes, helping users examine:- Mutation burden per gene
- Mutation impact distribution
- How mutations are distributed across patients
- Multi-tool support for each top gene
It contains two interactive components.
Components
Mutation Impact Distribution of Top 30 Driver Genes (Left Plot)The plot displays mutation data across the top 30 driver genes, with each row representing a different driver gene and each column representing an individual patient or sample. Each cell within the plot indicates whether that particular sample carries a mutation in the corresponding gene, and if so, the predicted impact level of that mutation—categorized as either high impact, moderate impact, or low impact.
Additional Elements:
- Left panel (A): total mutation percentage per gene.
- Top bar chart (B): total mutation count per patient.
- Right bar chart (C): total mutation count per gene
Tool Support for Top 30 Driver Genes (Right Plot)
The plot displays a bar chart where each bar represents a gene, with the height of the bar indicating the number of mutation tools that identified that gene as a mutation driver. Genes that are supported by a greater number of tools suggest higher-confidence driver roles, as consensus across multiple computational methods provides stronger evidence for their functional importance in cancer development.
1.6 Cancer CNV
1.6.1 Overview
The Cancer CNV section visualizes genes exhibiting significant copy number variation (CNV) gain or loss in the selected cancer type. It summarizes CNV driver evidence across patient samples, chromosomes, and pathway enrichments, helping users explore CNV–expression relationships and CNV-driven biological mechanisms.
At the top of the tab, users may choose between two CNV driver–detection modes:- iGC (single-tool mode): displays CNV drivers predicted by the iGC algorithm.
- iGC ∩ DIGGIT (two-tool intersection mode): displays only genes identified as CNV drivers by both iGC and DIGGIT, providing a more stringent, consensus-based driver set.

Switching between modes allows users to compare tool-specific vs multi-tool consensus CNV drivers.
This section contains three components:- Visualization of Top 30 CNV Driver Genes
- Locus Enrichment
- CNV Driver Gene Summary Table
1.6.2 Visualization of Top 30 CNV Driver Genes
This panel presents CNV gain, loss, and neutral patterns for the top 30 CNV driver genes in the selected cancer type.
CNV Gain and Loss Distribution of Top 30 Genes (Top Chart)
The plot displays a bar chart summarizing the percentage of samples exhibiting copy number variation (CNV) changes across the top 30 CNV driver genes, with each bar color-coded to show CNV gain (pink), CNV loss (green), and no CNV change (blue). Users can hover over any bar segment to view the exact percentages of gain, loss, and neutral CNV states for each gene. Genes with high gain percentages may represent potential oncogenes, while those with high loss percentages may be tumor suppressor candidates, whereas genes with balanced or low CNV changes may indicate lower CNV-driven relevance in cancer development.
CNV Patterns of Top 30 Genes Across Cancer Samples (Bottom Heatmap)
The heatmap displays copy number variation (CNV) data with rows representing the top 30 CNV driver genes and columns representing individual patient samples, where each cell is color-coded to indicate CNV gain (pink), CNV loss (green), or no CNV event (blue). Additional summary panels provide complementary information: the left panel (A) shows total CNV gain/loss percentages per gene, the top bar chart (B) displays total CNV events per sample, and the right bar chart (C) presents total CNV events per gene. Rows dominated by green or pink indicate consistent CNV-driven alterations in specific genes, while samples with tall bars in the top chart may represent CNV-heavy tumor genomes, and genes showing both high CNV frequency and strong tool support from the summary table emerge as strong CNV driver candidates.
1.6.3 Locus Enrichment
This section explores chromosomal distribution and functional enrichment of CNV-associated genes.
Chromosomal Locus Enrichment of CNV-Associated Genes (Left Plot)
The plot displays each gene as a red dot positioned according to its chromosomal coordinates across the genome, with hovering over any dot revealing detailed information including the chromosome, position, gene symbol, and correlation value between CNV and expression. Dense clusters of dots indicate chromosomal regions enriched for CNV events, while genes showing high CNV–expression correlation may reflect dosage-sensitive drivers where copy number changes directly influence gene expression levels and potentially contribute to cancer development.
Locus Enrichment Summary Table (Right Table)
The table displays pathways or functional categories that are enriched among CNV-affected genes, helping users identify biological processes potentially disrupted by copy number variation events. Enrichment of pathways such as cell cycle regulation, DNA repair, or receptor tyrosine kinase (RTK) signaling may highlight key CNV-driven mechanisms underlying cancer development and progression.
1.6.4 CNV Driver Gene Summary Table
This table provides gene-level CNV statistics, including significance metrics, sample proportions, CNV amplitude, and CNV–expression associations, offering a comprehensive overview of copy number variation patterns across genes. Genes with significant gain or loss (low p-value or FDR) and high sample proportions represent strong CNV candidates, while positive CNV–expression correlations indicate copy-number–driven expression changes where genomic alterations directly influence gene expression levels. Combining this table with the heatmap helps confirm consistent CNV patterns across patients and strengthens the evidence for identifying clinically relevant CNV-driven genes.
1.7 Cancer Methylation
1.7.1 Overview
The Cancer Methylation section visualizes genes exhibiting significant hypermethylation or hypomethylation in the selected cancer type. It provides an overview of methylation driver distributions across patient samples and chromosomal locations, helping users explore methylation–expression relationships and identify epigenetically driven gene dysregulation.
At the top of the tab, users may select results generated by:- MethylMix alone (single-tool mode)
- The intersection of MethylMix and ELMER (two-tool mode)
 The intersection mode highlights high-confidence methylation drivers supported by both algorithms.
The intersection mode highlights high-confidence methylation drivers supported by both algorithms.
This section contains three components:
- Visualization of Top 30 Methylation Driver Genes
- Locus Enrichment
- Methylation Driver Gene Summary Table
1.7.2 Visualization of Top 30 Methylation Driver Genes
This panel summarizes the methylation status of the top 30 methylation driver genes and shows how hypermethylation/hypomethylation patterns appear across samples.
Methylation Status of Top 30 Genes (Top Bar Chart)
The plot displays a bar chart summarizing the proportion of samples showing hypermethylation (pink), hypomethylation (green), and no methylation change (blue) across the top 30 methylation driver genes, with each bar representing a single gene. Users can hover over any bar to view the exact percentages of hyper-, hypo-, and unmethylated samples for that gene. Genes with high hypermethylation may involve promoter silencing or epigenetic downregulation that reduces gene expression, while genes with high hypomethylation may indicate enhancer activation or derepression leading to increased expression, whereas balanced patterns may suggest context-specific or mixed methylation states that vary across different tumor samples or subtypes.
Methylation Patterns Across Cancer Samples (Bottom Heatmap)
The heatmap displays methylation data with rows representing the top 30 methylation driver genes and columns representing individual patient samples, where cell colors indicate hypermethylation (pink) or hypomethylation (green). Additional summary bars provide complementary information: the left panel (A) shows total methylation percentage per gene, the top bar chart (B) displays total methylation events per sample, and the right bar chart (C) presents total methylation events per gene. Genes with predominantly pink rows are consistently hypermethylated across patients, while those with predominantly green rows show consistent hypomethylation, and tall bars in the top chart indicate samples with high methylation burden. Comparison with expression data through correlation analysis helps identify methylation-driven expression changes, revealing epigenetic mechanisms that influence gene activity in cancer.
1.7.3 Locus Enrichment
This section maps methylation-associated genes to their chromosomal positions and evaluates pathway enrichment.
Chromosomal Locus Enrichment of Methylation-Associated Genes (Left Plot)
The plot displays each methylation-associated gene as a red dot positioned according to its chromosomal coordinates across the genome, with hovering over any dot revealing the chromosome, genomic position, gene symbol, and correlation value between methylation and expression. Clusters of dots may indicate epigenetically altered chromosomal regions where multiple genes experience coordinated methylation changes, while positive correlation values suggest that methylation changes strongly influence gene expression, such as hypermethylation leading to downregulation or hypomethylation resulting in upregulation. Genes with high correlation values may represent functional methylation drivers where epigenetic modifications play a critical role in regulating gene activity and contributing to cancer phenotypes.
Locus Enrichment Summary Table (Right Table)
The table displays pathways enriched among methylation-affected genes, helping users identify biological processes impacted by epigenomic dysregulation in cancer. Enrichment in pathways such as DNA repair, immune regulation, or cell differentiation may indicate core mechanisms that are altered via methylation changes, revealing how epigenetic modifications contribute to tumor development, progression, and immune evasion.
1.7.4 Methylation Driver Gene Summary Table
This table summarizes methylation statistics for each gene identified as a methylation driver in the selected cancer type, integrating results from MethylMix and ELMER outputs and including methylation proportions, probe-level data, and correlations with gene expression. Strong hyper- or hypomethylation percentages combined with significant adjusted p-values indicate robust methylation drivers, while a strong negative correlation typically suggests promoter hypermethylation reducing expression and a positive correlation may indicate intragenic methylation effects that enhance gene activity. Genes showing consistent patterns across both tools (MethylMix ∩ ELMER) represent high-confidence methylation drivers, as convergent evidence from multiple computational methods strengthens the reliability of epigenetic alterations as key regulatory mechanisms in cancer.
1.8 Cancer Survival
1.8.1 Overview
The Cancer Survival section visualizes the survival relevance and synergistic effects of driver genes in the selected cancer type. By integrating survival outcomes with gene expression profiles, this section identifies gene pairs whose combined expression patterns have a stronger impact on patient prognosis than either gene alone.
The section contains two components:- Survival Synergy Network
- Survival of Synergistic Effect
1.8.2 Survival Synergy Network
Purpose
The Survival Synergy Network illustrates synergistic survival relationships between genes identified as survival-relevant in the selected cancer.
A synergistic effect occurs when the joint hazard ratio (HR) of two genes exceeds each gene’s individual HR, suggesting their combined expression amplifies risk (HR > 1) or provides enhanced protection (HR < 1).
- 1.5-fold synergy
- 2-fold synergy
- Positive direction (HR > 1) → worse prognosis
- Negative direction (HR < 1) → better prognosis
Gene pairs with opposite HR directions (e.g., one HR > 1 and one HR < 1) are excluded because they cannot form meaningful synergy.
Nodes
- Represent individual survival-relevant genes
- Can be filtered by Gene Source:
- CGC (Cancer Gene Census)
- NCG6.0 (Network of Cancer Genes)
- All genes
Edges
- Connect gene pairs with validated synergistic survival effects
- Edge presence indicates that the joint HR exceeds each single-gene HR by ≥1.5× or ≥2×
Interaction Guide
Users can explore the network using several interactive options:Node Interaction
- Click a node to highlight its connected synergistic partners
- Click on blank space to reset and return to the full network
- Use the dropdown to directly select and highlight a specific gene
- Gene Source: CGC, NCG, or All
- Synergistic Effect Level: 1.5× or 2×
- HR Direction:
- >1 (synergy associated with poor prognosis)
- <1 (synergy associated with favorable prognosis)
- Both

1.8.3 Survival of Synergistic Effect
Purpose
This section provides detailed survival analyses for each synergistic gene pair identified in the network, containing a table of synergistic gene pairs with associated statistics and survival plots illustrating how combined gene expression affects patient prognosis.
Synergistic Gene-Pair Table (Left Panel)
The table displays survival statistics for each gene pair, listing the cancer type, Gene 1 and Gene 2 identifiers, HR fold change comparing All.high versus Others, log-rank p-values, 4-group log-rank p-values, and Log2HR values for three comparisons: All.high vs All.low, High.low vs All.low, and Low.high vs All.low. Users can select an HR direction (>1 or <1), choose a specific gene pair from the table, and view the corresponding survival plots on the right panel. Only gene pairs where the joint hazard ratio meaningfully exceeds individual HRs are included, and all p-values in the table meet significance thresholds (p < 0.05), ensuring robust synergistic interactions.
Survival Plots (Right Panel)
Four-Group Survival PlotThe four-group survival plot displays survival outcomes for all combinations of gene expression: All.high (purple, both genes high), All.low (red, both genes low), Low.high (green, gene1 low and gene2 high), and High.low (blue, gene1 high and gene2 low), where distinct separation between curves indicates strong synergistic or antagonistic effects and differences in survival trends help identify which expression groups drive the synergy. The two-group comparison plot contrasts the All.high group against all other combinations combined, where a significantly worse outcome for All.high suggests a synergistic negative effect on survival while a significantly better outcome indicates a synergistic protective effect.
Interpretation
HR values reflect the risk magnitude associated with each expression group, while log-rank p-values indicate statistical significance of survival differences between groups. Users can hover over survival curves to view time in months and survival probability at specific points, and combined interpretation of both plots helps evaluate both the overall combinatorial effects and the specific expression groups driving synergistic interactions in cancer outcomes.
1.9 Cancer miRNA
1.9.1 Overview
The Cancer miRNA section visualizes regulatory relationships between differentially expressed (DE) genes and miRNAs in the selected cancer type. It integrates both experimentally validated interactions and computationally predicted miRNA–target relationships to help users identify miRNA regulators, target genes, and expression patterns associated with carcinogenesis.
This section contains three components:- miRNA–Gene Interaction Network
- Visualization of Differentially Expressed Genes and miRNAs
- Gene–miRNA Correlation Summary Table
1.9.2 miRNA-Gene Interaction Network
Purpose
This interactive network displays validated and predicted interactions between genes and miRNAs, enabling users to explore regulatory mechanisms that may contribute to cancer development or progression.
Nodes
- Gene nodes represent DE or driver-relevant genes
- miRNA nodes represent DE miRNAs or miRNAs predicted/validated to regulate those genes
Edges
Two types of miRNA–gene interactions are shown:- Validated interactions
- Experimentally supported miRNA–target interactions
- Sourced from miRTarBase, where:
- 1 = supported by at least one experimental study
- 2 = supported by multiple independent studies or experimental methods
- Visible only when the 'Validated' checkbox is checked
- Predicted interactions
- Derived from 12 bioinformatics prediction tools, including: DIANA-microT, miRDB, TargetScan, RNAhybrid, miRanda, PITA, PicTar, RNA22, and others
- Users may set a minimum prediction support threshold (≥6, ≥8, or ≥10 tools)
Note: All interactions appear as dotted lines in the visualization. The distinction between validated and predicted interactions is determined by whether the 'Validated' checkbox is enabled, not by line style. Edges grow denser as evidence increases (validated + multi-tool predictions).
Interaction Guide Selecting Nodes
- Click a node to highlight all connected partners
- Click blank/white space to reset the full network
- Use the dropdown to directly locate and highlight a specific gene
- Gene Source: CGC, NCG, or All
- Minimum Prediction Support: ≥6, ≥8, or ≥10 prediction tools
- Validation Status: Show or hide validated interactions
- High-confidence regulatory interactions
- Oncogenic or tumor suppressive miRNA–gene pairs
- Experimentally supported vs computationally predicted relationships

1.9.3 Visualization of Differentially Expressed Genes and miRNAs
Purpose
This heatmap displays the expression profiles of differentially expressed (DE) genes and DE miRNAs across tumor and normal samples, allowing users to compare regulatory patterns at the expression level and examine relationships between miRNA regulators and their target genes.
Heatmap Display
The heatmap presents rows representing DE miRNAs and/or DE genes and columns representing individual patient samples, with a color scale where red indicates higher expression and blue indicates lower expression. Sample labels distinguish between TP (dark blue, tumor samples) and NT (light blue, normal samples), while clustering dendrograms show similarities among samples (top) and similarities among genes/miRNAs (right), revealing co-expression patterns and sample groupings.
Visualization Modes
Users can switch between three visualization modes: DE miRNA (displaying only DE miRNAs), DE gene (displaying only DE genes), or DE miRNA + DE gene (combined view). These modes help users examine upregulated miRNAs versus their target genes, identify opposing expression trends such as miRNA upregulation with corresponding gene downregulation, and detect co-expression clusters among miRNAs or genes that suggest coordinated regulatory mechanisms.
Interpretation Tips
Inverse expression patterns, where miRNA expression is high and target gene expression is low, may indicate miRNA-mediated repression as a functional regulatory mechanism. Co-clustering of genes or miRNAs suggests shared regulatory pathways or common biological functions, while DE miRNAs that align with network interactions identified in other analyses highlight strong regulatory candidates with potential functional significance in cancer development or progression.
1.9.4. Gene–miRNA Correlation Summary Table
This table provides quantitative measures of gene–miRNA regulatory relationships using correlation analysis, validation data, and prediction support to assess the strength and reliability of regulatory interactions. Negative correlations often indicate miRNA-mediated repression where miRNA upregulation corresponds with target gene downregulation, while positive correlations may suggest co-regulation or indirect regulatory mechanisms involving intermediate factors. High prediction-tool support combined with validated status and strong correlation values indicate high-confidence interactions that are likely functionally relevant, and users can cross-check this table with network edges and heatmap expression patterns to confirm consistent regulatory relationships across multiple analytical approaches.
1.10 Cancer Multi-omics
1.10.1 Overview
The Cancer Multi-Omics section visualizes driver genes identified through multi-omics integration tools in the selected cancer type. By combining evidence across mutations, CNV, methylation, mRNA expression, and miRNA regulation, this section highlights genes supported by multiple molecular layers and explores their biological functions, tool support, and distribution across omics categories.
Users may analyze:- All genes
- Only genes included in Cancer Gene Census (CGC)
- Network of Cancer Genes (NCG6.0)
 The section contains three components:
The section contains three components:
- Multi-Layer Relationship Diagram of Multi-Omics Drivers and Biological Functions
- Distribution of Multi-Omics Drivers Across Omics Layers
- Cross-Tool Comparison of Multi-Omics Driver Detection
1.10.2 Multi-Layer Relationship Diagram of Multi-Omics Drivers and Biological Functions
This section presents a diagram illustrating hierarchical relationships from the cancer type → omics layers → multi-omics driver genes → Gene Ontology (GO) functions, showing how integrative driver events connect molecular alterations to biological processes. A summary table below the diagram lists detailed gene-specific and GO-specific results.
1.10.3 Distribution of Multi-Omics Drivers Across Omics Layers
Purpose
This section summarizes how many tools identify each gene as a multi-omics driver and how those drivers are distributed across omics categories.
It consists of two complementary plots:- Left Heatmap – Tool Support per Gene and Omics Layer
- Right Bar Chart – Top Genes by Multi-Omics Tool Support
Tool Support Across Omics Layers (Left Heatmap)
This heatmap displays tool support across omics layers, with rows representing multi-omics driver genes, columns representing different omics layers, and cells showing the number of tools that identified each gene within that specific omic layer, where hovering over any cell reveals the exact number of supporting tools. Darker cells indicate stronger multi-tool evidence for that particular omic layer, suggesting robust detection across computational methods, while genes with support across multiple omics layers may represent high-confidence integrative drivers that are dysregulated through multiple molecular mechanisms. Missing or light-colored cells suggest omics-specific drivers where the gene shows alterations predominantly in one molecular layer rather than across multiple platforms.
Top Genes by Support Count (Right Bar Chart)
This bar chart displays an ordered list of genes ranked by the total number of supporting tools, with the x-axis showing tool counts and the y-axis displaying gene symbols, where bar colors differentiate the omics categories contributing to each gene's overall support. Users can hover over any bar to view the number of tools per omic layer contributing to each gene's score, providing detailed breakdowns of evidence sources. Genes with the longest bars are most consistently supported across computational tools and represent the strongest driver candidates, while multicolored bars indicate multi-layered evidence across different omics platforms suggesting integrative dysregulation, and single-color bars represent omics-specific drivers that show alterations predominantly within one molecular layer.

1.10.4 Cross-Tool Comparison of Multi-Omics Driver Detection
Purpose
This section compares the coverage and consistency of different multi-omics driver-identification tools across omics layers.
It contains:- Left Heatmap – Tool vs. Omics Layer Coverage
- Right Bar Chart – Gene Counts by Tool Support Level
Proportion of Genes Identified by Each Tool (Left Heatmap)
This heatmap displays multi-omics identification tools on the y-axis and omics layers on the x-axis, with each cell showing the proportion of genes identified by a specific tool for a given omic layer, where hovering reveals exact proportion values. High-proportion cells reveal tool specialization or sensitivity toward certain omics categories, indicating that some tools are particularly effective at detecting drivers within specific molecular layers, while tools with balanced proportions across multiple omics may provide more integrative coverage and capture dysregulation across diverse biological mechanisms.
Gene Counts by Tool Support Level (Right Bar Chart)
This bar chart displays the distribution of tool support across genes, with the x-axis showing the number of tools supporting a gene and the y-axis showing the number of genes at each support level, where hovering reveals the exact count of genes supported by each tool count. A right-skewed distribution, where more genes are supported by many tools, indicates strong cross-tool consensus and robust identification of driver genes across computational methods, while a left-skewed distribution suggests tool divergence where few genes are consistently detected across platforms. Genes supported by more tools typically represent high-confidence multi-omics drivers, as convergent evidence from multiple analytical approaches strengthens their credibility as functionally relevant cancer-associated genes.

2. Gene
2.1 Gene Module Overview
The Gene module provides a comprehensive, multi-omics overview of a user-selected gene across multiple cancer types. By integrating information from expression, mutation, CNV, methylation, survival relevance, miRNA regulation, protein expression, and multi-omics driver evidence, this module helps users understand how a gene behaves across the cancer landscape.
2.2 Input Selection
To begin, choose how you want to search for the gene:Search Mode
- Gene Name – Enter the official HGNC gene symbol (e.g., TP53)
- Ensembl ID – Enter the Ensembl gene identifier (e.g., ENSG00000141510)

After entering the gene, click Submit to generate all downstream results.
2.3 Overview of Result Tabs
The Gene module contains several results tabs, each summarizing multi-omics evidence and functional insights for a selected gene across different cancer types:- Summary – Integrates cross-omics alterations and clinical associations to provide an overview of the gene's role, including mutation frequency, expression patterns, and survival impact across cancers.
- Expression – Displays differential expression profiles of the gene across tumor versus normal samples, with tissue-specific patterns and expression correlations.
- Mutation – Catalogs mutation types, hotspots, and functional consequences, including pathogenicity predictions and mutation distribution along protein domains.
- CNV – Visualizes copy-number alterations affecting the gene, including amplification and deletion frequencies across cancer types and CNV–expression correlations.
- Methylation – Highlights methylation status at the gene locus, including promoter hyper- or hypomethylation patterns and methylation–expression relationships.
- Survival – Presents survival analysis based on gene expression, mutation status, or other alterations, identifying prognostic significance across different cancer cohorts.
- miRNA – Shows regulatory interactions between the gene and miRNAs, including experimentally validated and predicted miRNA–target relationships.
- Protein – Displays protein-level expression data, post-translational modifications, and protein–protein interactions relevant to the gene product.
- Multi-Omics – Integrates evidence across all omics layers to reveal comprehensive functional patterns, cross-omics correlations, and the gene's role in oncogenic processes.
2.4 Gene Summary
Overview
The Gene Summary tab presents an integrated Gene Overview Heatmap, summarizing key molecular features of the selected gene across cancer types. It helps users quickly identify whether a gene is dysregulated, mutated, copy-number altered, methylated, survival-associated, miRNA-regulated, or supported by multi-omics driver tools.
Results are displayed across TCGA datasets (left heatmap) and non-TCGA datasets (right heatmap).Asterisks (*) mark cancer types where the gene is identified as a multi-omics driver.

Heatmap Features
Each column represents an omics feature. Colors and intensities communicate the strength or direction of the molecular signal.
Below is the standardized interpretation for each heatmap row:- Differential Expression (DE): This layer indicates whether the gene is significantly differentially expressed (p < 0.05), with red representing upregulated genes (log₂FC > 1) and green representing downregulated genes (log₂FC < –1), where darker colors indicate stronger fold changes. This helps identify whether the gene is consistently upregulated or downregulated in specific cancer types.
- Mutation: This layer represents the number of computational tools identifying the gene as a mutation driver, with blue shades indicating increasing tool support where darker blue corresponds to more tools. Genes with dark-blue cells are consistently predicted as mutation drivers across multiple computational methods, suggesting high-confidence functional mutations.
- Copy Number Variation (CNV): This layer shows whether the gene exhibits CNV gain or loss, with red indicating copy number gain (1) and green indicating copy number loss (–1). This helps distinguish oncogene-like patterns characterized by copy number gains from tumor suppressor-like patterns characterized by copy number losses.
- Methylation: This layer indicates whether the gene is hyper- or hypomethylated, with red representing hypermethylation (1) and green representing hypomethylation (–1). Consistent methylation patterns across cancer types may indicate epigenetic regulation of gene expression that influences cancer development or progression.
- Survival: This layer shows survival relevance based on log-rank test (p < 0.05), with red indicating oncogene-like behavior (log₂HR > 0) and green indicating tumor suppressor-like behavior (log₂HR < 0), where darker colors represent stronger associations with survival. This highlights whether high expression of the gene predicts worse or better patient outcomes.
- miRNA Regulation: This layer displays the number of miRNAs predicted or validated to interact with the gene, with orange shades indicating increasing numbers of miRNA interactions where darker orange corresponds to more interactions. Genes with many interacting miRNAs may be heavily regulated at the post-transcriptional level, suggesting complex regulatory control mechanisms.
2.5 Gene Expression
2.5.1 Overview
The Gene Expression module visualizes transcript abundance (TPM) of the selected gene across cancers. Users can explore how gene expression varies across different biological categories—including sample types, tumor vs. normal tissues, mutation classes, and tumor stages—with both pan-cancer and cancer-specific visualizations.
Each tab includes:
- Pan-Cancer View — expression distributions across all cancer types
- Cancer-Specific View — detailed expression profiles within a selected cancer type
- Toggle groups using the legend
- Hover over points for sample-level metadata (cancer type, tissue, grouping variable, TPM)
- Hover near box/violin areas to obtain summary statistics
(max, upper fence, Q3, median, Q1, lower fence, min)
2.5.2 Expression by Sample Type (All Categories)
This tab displays expression across all available sample types (e.g., NT, TP, TM, TRB, TBM).
Pan-Cancer View: Expression by Sample Type (All Categories)
Boxplots show TPM expression of the selected gene across all cancer types (x-axis), grouped and colored by sample type.Users can:
- Toggle sample-type groups
- Hover for sample metadata
- Hover near box areas for distribution summaries
- NT — Solid Tissue Normal
- TP — Primary Solid Tumor
- TM — Metastatic
- TR — Recurrent Solid Tumor
- TAM — Additional Metastatic
- TAP — Additional New Primary
- TB — Primary Blood-Derived Cancer
- TBM — Metastatic Blood-Derived Cancer
- TRB — Recurrent Blood-Derived Cancer
- TRBM — Recurrent Blood-Derived Metastatic
- Tumor-normal contrasts may reveal dysregulated expression patterns.
- Differences across sample subclasses may indicate metastasis-, blood-derived-, or recurrence-associated changes.

Cancer-Specific View: Expression by Sample Type (All Categories)
The cancer-specific view displays violin plots showing log₁₀(TPM) expression values for the selected cancer type, grouped by sample type, with a left panel for selecting the cancer type and a right panel displaying the violin plot by sample type, accompanied by a bottom summary table containing p-values comparing groups. Users can toggle sample-type groups on or off, hover over individual data points to view sample metadata, and hover on violin shapes to see distribution statistics such as median and quartile values. Significant p-values indicate meaningful expression differences among sample-type groups, and clear separation between NT (normal tissue) and TP (primary tumor) samples may suggest diagnostic relevance where the gene could serve as a potential biomarker for distinguishing tumor from normal tissue.
2.5.3 Expression by Sample Type (Normal vs. Tumor)
This tab isolates the contrast between Normal (NT) and Primary Tumor (TP) samples.
Pan-Cancer View: Expression by Sample Type (Normal vs. Tumor)
Boxplots display TPM expression across cancer types for NT and TP samples only.Users can:
- Toggle NT and TP groups
- Hover for metadata and boxplot summaries
- TP > NT: potential oncogenic upregulation
- TP < NT: potential tumor suppressor downregulation

Cancer-Specific View: Expression by Sample Type (Normal vs. Tumor)
The cancer-specific view displays violin plots showing log₁₀(TPM) expression values for the selected cancer type, grouped by sample type, with a left panel for selecting the cancer type and a right panel displaying the violin plot by sample type, accompanied by a bottom summary table containing p-values comparing groups. Users can toggle sample-type groups on or off, hover over individual data points to view sample metadata, and hover on violin shapes to see distribution statistics such as median and quartile values. Significant p-values indicate meaningful expression differences among sample-type groups, and clear separation between NT (normal tissue) and TP (primary tumor) samples may suggest diagnostic relevance where the gene could serve as a potential biomarker for distinguishing tumor from normal tissue.
2.5.4 Expression by Mutation Class
This tab evaluates expression levels across mutation impact categories, including:- High
- Moderate
- Low
- Modifier
- Normal Tissue
- Tumors without Mutation
Pan-Cancer View: Expression by Mutation Class
Boxplots display TPM expression across cancer types, grouped by mutation class.
User can:- Toggle mutation classes
- Hover for sample metadata
- Hover near box areas for summary statistics
- Expression differences between “High Impact” vs “No Mutation” may indicate mutation-driven regulatory effects.

Cancer-Specific View: Expression by Mutation Class
Violin plots display log₁₀(TPM) expression values for the selected cancer type, grouped by mutation class, where users can select the cancer type in the left panel and view the corresponding expression distributions across different mutation classes. The summary table provides pairwise p-values comparing mutation classes and sample counts per class, allowing users to assess the statistical significance and sample sizes underlying each comparison. Mutation-class differences reflect the gene's sensitivity to mutational context, and significant separation between classes suggests mutation-dependent transcriptional changes where the presence or type of mutation influences gene expression levels.
2.5.5 Expression by Tumor Stage
This tab examines expression variation across clinical tumor stages (I–IV).
Pan-Cancer View: Expression by Tumor Stage
Boxplots show TPM expression across cancer types, grouped by tumor stage.Users can:
- Toggle stages
- Hover for metadata and distribution summaries
- Stage-associated patterns may reflect biological roles in progression or severity.

Cancer-Specific View: Expression by Tumor Stage
Violin plots display log₁₀(TPM) expression values for the selected cancer type, grouped by tumor stage, where users can select the cancer type in the left panel and view expression distributions across different disease stages. The summary table includes p-values for stage comparisons and sample counts per stage, providing statistical support and sample size information for each stage group. Consistent expression changes across stages suggest the gene's involvement in cancer progression, where increasing or decreasing expression trends may indicate the gene's role in tumor advancement from early to advanced disease stages.

2.6 Gene Mutation
2.6.1 Overview
The Gene Mutation module visualizes mutation patterns of the selected gene across cancer types.Results incorporate mutation calls, mutation impact categories, and mutation driver predictions from multiple mutation-identification tools.
Users can explore:
- Mutation rates and percentages along protein coordinates
- Mutation hotspots within each cancer
- Exon-level mutation distributions
- Alignment of mutations with protein domains
- Summary mutation statistics
2.6.2 Mutation Rate
Pan-Cancer View: Mutation Rate Heatmap
This view integrates multiple coordinated panels to show where mutations occur along the protein and how frequently they appear across cancer types, using mutation rate as the metric.
ComponentsA. Pan-Cancer Mutation Hotspot Heatmap
This heatmap displays cancer types as rows and protein positions as columns, with cell color indicating the mutation rate at each specific position. Users can hover over cells to view the cancer type, protein position, and mutation rate, allowing them to identify protein regions with recurrent mutation hotspots across multiple cancer types.
B. Protein Region Impact Bar PlotThis plot aggregates mutation rates per protein region, with bars stacked by impact level (High, Moderate, or Low) to show the relative contribution of different mutation severities. Hovering over bar segments reveals the region name, impact category, and mutation rate, demonstrating which functional regions of the protein accumulate the highest mutation load.
C. Dataset-Level Mutation Burden Bar PlotThis bar chart displays the overall mutation rate per dataset or cancer type, with bars stacked by mutation impact level to show the distribution of mutation severities. Users can hover to view the dataset, tissue type, impact level, and mutation rate, providing a quick comparison of which cancers have the heaviest mutation burden for the selected gene.
D. Dataset & Tissue LegendThis companion panel lists the tissue type, project ID, and cancer type for each dataset included in the analysis, with each color corresponding to a specific tissue type to help users interpret the color-coding used throughout the visualization.
E. Protein Domain Annotation TrackThis track shows annotated protein domains from Pfam or InterPro databases, displaying the domain name, protein coordinate range, and functional description (accessible via hover). This annotation aligns functional domains with mutation hotspots, helping users understand whether mutations cluster in functionally important regions of the protein.
F. Exon Annotation TrackThis track displays exon boundaries aligned to protein coordinates, with each exon shown as a distinct colored block to illustrate the genomic structure underlying the protein sequence and how mutations map to specific exons.
G&H. LegendsTwo legends accompany the visualization: a mutation rate legend providing a continuous color scale for heatmap intensity, and an impact legend showing categorical colors for High, Moderate, and Low mutation impacts to help users interpret the color-coding throughout all components.

Cancer-Specific View: Mutation Rate Bar Chart
This visualization shows mutation rate across protein positions for the selected cancer type, allowing users to examine cancer-specific mutation patterns along the protein sequence. The left panel (A) enables users to select a cancer type, while the right panel (B) displays a bar chart of mutation rate along the protein, with bars stacked by impact level (High, Moderate, and Low) to show the distribution of mutation severities at each position.
Users can hover over any bar to view detailed information including the protein position, mutation rate, impact category, and mutation count for that specific location. An impact legend (C) provides the color key for interpreting High, Moderate, and Low mutation impacts throughout the visualization. This analysis helps identify driver-related hotspot regions specific to a given cancer type, revealing which protein positions are preferentially mutated in particular cancer contexts and potentially highlighting functionally important sites for targeted therapeutic development.
2.6.3 Mutation Percent
Pan-Cancer View: Mutation Percent Heatmap
This visualization is structurally identical to the Mutation Rate view but uses mutation percentage—the proportion of mutated samples in each dataset—rather than mutation rate.
ComponentsA. Pan-Cancer Mutation Hotspot Heatmap
This heatmap displays cancer types as rows and protein positions as columns, with cell color indicating the mutation percentage at each specific position. Users can hover over cells to view the cancer type, protein position, and mutation percentage, revealing which protein positions are most frequently mutated across patient samples in different cancer types.
B. Protein Region Impact Bar Plot
This plot shows mutation percentage per protein region, with bars stacked by mutation impact level to display the relative contribution of High, Moderate, and Low impact mutations. Hovering over bar segments reveals the region name, impact level, and mutation percentage, highlighting protein regions with high prevalence of mutations among patients and indicating which functional domains are most commonly affected.
C. Dataset-Level Mutation Burden Bar Plot
This bar chart displays mutation percentage per dataset or cancer type, with bars stacked by impact level to show the distribution of mutation severities. Users can hover to view the dataset, tissue type, impact level, and mutation percentage, identifying cancer types where mutations in the gene are widespread across patient populations.
D. Dataset & Tissue Legend
Color-coded tissue and dataset identifiers for interpreting the heatmap rows.
E. Protein Domain Annotation Track
This track displays Pfam and InterPro protein domains aligned to protein coordinates, showing the domain name, coordinate range, and functional details accessible through hovering, allowing users to determine whether mutations cluster within functionally important protein domains.
F. Exon Annotation Track
This track displays exon boundaries aligned to protein structure, with each exon shown as a distinct block to illustrate how the genomic organization corresponds to the protein sequence and mutation positions.
G&H. Legends
Two legends accompany the visualization: a mutation percent legend providing a color scale for mutation proportions, and an impact legend showing colors for High, Moderate, and Low mutation impact categories to help users interpret the color-coding throughout all components.

Cancer-Specific View: Mutation Percent Bar Chart
This visualization shows the percentage of samples with mutations at each protein coordinate for the selected cancer type, allowing users to examine mutation prevalence across patient populations. The left panel (A) enables users to select a cancer type, while the right panel (B) displays a bar chart of mutation percentage along the protein, with bars stacked by impact level to show the distribution of mutation severities at each position. An impact legend (C) provides the color key for interpreting High, Moderate, and Low mutation impacts throughout the visualization. Users can hover over any bar to reveal the protein region, percentage of affected samples, and mutation impact level. This analysis is useful for understanding mutation prevalence rather than mutation density, helping identify which protein positions are mutated in a significant proportion of patients and may represent clinically relevant alterations for diagnostic or therapeutic targeting.
2.6.4 Exon Distribution
Pan-Cancer View: Exon Mutation Distribution
ComponentsA. Mutation Count by Exon
Shows the number of mutations per exon across all cancer types. X-axis = exon number; Y-axis = mutation count. Bars are stacked by mutation impact (High, Moderate, Low, Modifier). Hover to view exon number, impact, and mutation count.
B. Mutation Percentage by Exon
Displays the proportion of mutated samples per exon. X-axis = exon number; Y-axis = mutation percentage. Hover to view exon number, impact, and mutation percentage.
C. Protein Domain Panel
Annotated functional domains with Pfam ID, InterPro ID, position, and description. Hover for details on each domain.
D. Exon Annotation Track
Each colored block represents an exon aligned to the protein coordinate axis.
E. Impact Legend

Cancer-Specific View: Exon Mutation Distribution
This visualization provides controls for users to customize their analysis, including a 'Visualization By' option (A) to switch between mutation count or mutation percentage, a cancer type panel (A) to select a specific cancer, and a legend (D) to hide or show impact categories for clearer viewing. The right panel (B) displays a bar plot showing mutation distribution across exons, with bars color-coded by impact level (High, Moderate, Low) to visualize the severity of mutations within each exon. Users can hover over any bar element (C) to view detailed information including the exon number, mutation count or percentage, impact level, and cancer type. This analysis reveals exon-specific mutation hotspots within a selected cancer, helping identify which exons of the gene accumulate the most mutations and may harbor functionally critical alterations that drive cancer development or progression in specific cancer contexts.
2.7 Gene CNV
2.7.1 Overview
The Gene CNV module visualizes copy-number variation (CNV) patterns of the selected gene across multiple cancer types.
Results integrate predictions from two CNV-identification tools — iGC and DIGGIT — and provide:- A pan-cancer overview of CNV gain/loss significance
- Detailed cancer-specific CNV–expression correlations
- A summary table with statistical and tool-specific CNV information
Users can explore how CNV gain or loss relates to expression changes and identify cancers where the gene behaves as a potential CNV driver.
This module includes three sections:- Pan-Cancer View: Copy Number Variation Overview
- Cancer-Specific View: CNV Distribution and Correlation
- CNV Summary Table
2.7.2 Pan-Cancer View: Copy Number Variation Overview
This visualization shows CNV gain, loss, and neutral states for the selected gene across all cancer types, based on results from iGC and DIGGIT.
It consists of three main components that work together to display CNV patterns across cancer types. The CNV driver panel (A) at the top indicates how many CNV tools support the gene as a CNV driver in each cancer type, with light grey representing identification by iGC only and dark grey indicating identification by both iGC and DIGGIT, where darker shading suggests higher cross-tool confidence. The main panel (B) displays sample proportion bars for each cancer type, with bar segments color-coded to show the proportion of samples exhibiting CNV gain (red), CNV loss (green), or no CNV change (blue), where bar height reflects the percentage of affected samples. Users can hover over any bar (C) to view detailed information including the cancer type, CNV status (gain, loss, or none), tool support information (iGC, DIGGIT, or both), and proportional values. This view helps users determine which cancers show frequent CNV gains or losses, whether CNV patterns are supported by one or both CNV tools, and which cancer types may exhibit significant CNV-driven expression changes for the selected gene.
2.7.3 Cancer-Specific View: CNV Distribution and Correlation
This combined visualization examines how CNV states (gain, loss, none) affect expression levels within a specific cancer type. Users select a cancer dataset from the left panel, and the right side generates a multi-panel plot consisting of four interconnected components that together illustrate the relationship between copy number alterations and gene expression.
Components
A. CNV–Expression Correlation Scatter Plot (Upper-Right)
This scatter plot displays the relationship between segment mean (log₂(copy-number / 2)) on the x-axis and expression (TPM) on the y-axis, with each point representing an individual sample color-coded by CNV status: red for gain, blue for loss, green for none, and grey for normal. Users can hover over points to reveal exact segment mean values, expression levels, CNV status, and sample details. A positive correlation indicates that CNV gains associate with higher expression, while a negative correlation suggests that CNV losses associate with reduced expression, revealing whether copy number changes drive transcriptional alterations.
B. Expression by CNV Status Boxplot (Upper-Left)
This boxplot summarizes TPM expression across CNV categories (gain, loss, none), allowing users to compare expression distributions between groups. Hovering over boxplots reveals distribution metrics including maximum, upper fence, Q3, median, Q1, lower fence, and minimum values. This component highlights whether expression differs significantly across CNV groups, providing statistical evidence for CNV-mediated expression changes.
C. Segment Mean by CNV Status Boxplot(Bottom-Right)
This boxplot shows the distribution of segment means for each CNV status category, with hover details displaying segment mean ranges, quartiles, and fence limits. This visualization confirms whether gain/loss classifications are consistent with segment mean thresholds, validating the CNV categorization used throughout the analysis.
D. Correlation Summary (Bottom-Left)
This panel provides quantitative correlation metrics between segment mean and gene expression, displaying the Spearman correlation coefficient (ρ) and associated p-value. Strong absolute correlation values combined with significant p-values indicate CNV-driven expression regulation, where copy number alterations directly influence gene expression levels.
Segment Mean Reference
Segment mean is calculated as log₂(copy number / 2), with threshold interpretations as follows: values approximately 0 (−0.3 to 0.3) indicate diploid state with no CNV change, values greater than 0.3 indicate CNV gain, and values less than −0.3 indicate CNV loss.
Interactions
Users can hover over scatter plot points to view sample-level CNV and expression information, hover over boxplots to access full statistical summaries, and use the legend to toggle CNV states (gain, loss, none, normal) on or off. Selecting a new cancer type from the left panel automatically updates all four visualization components to reflect the chosen dataset.

2.7.4 CNV Summary Table
This table provides detailed tool-specific CNV statistics for the selected gene across cancer types, including correlation metrics that reveal the relationship between copy number alterations and gene expression. The Spearman correlation coefficient (ρ) indicates the direction and strength of this relationship: positive ρ values suggest that CNV gains are associated with higher expression levels, while negative ρ values indicate that CNV losses correspond to lower expression, and the accompanying p-value indicates the statistical significance of these correlations, helping users identify cancer types where CNV alterations functionally drive expression changes.
2.8 Gene Methylation
2.8.1 Overview
The Gene Methylation module visualizes DNA methylation patterns of the selected gene across cancer types and evaluates how methylation relates to gene expression.
Two complementary methylation tools are used:- MethylMix — identifies hypermethylated or hypomethylated genes based on β-value distributions
- ELMER — identifies functionally relevant methylation probes associated with gene expression changes
- Pan-Cancer View: Methylation Status Overview
- Cancer-Specific View: Methylation Distribution and Correlation
- Methylation Summary Table
2.8.2 Pan-Cancer View: Methylation Status Overview
This view summarizes methylation states of the selected gene across cancer types, integrating predictions from MethylMix (1) and ELMER (2).
It displays methylation patterns across cancer types through three integrated components. The methylation driver panel (A) at the top indicates whether each cancer type shows significant methylation-driven alteration and the level of tool support, with light grey representing identification by MethylMix only and dark grey indicating identification by both MethylMix and ELMER, where darker shading suggests stronger cross-tool evidence. The main display consists of sample proportion bars (B) for each cancer type, with bar segments color-coded to show the proportion of samples classified as hypermethylated (red), hypomethylated (green), or showing no methylation change (blue), where bar height represents the sample percentage within the cancer dataset. Users can hover over each bar (C) to reveal detailed information including the cancer type, methylation category, sample proportion, and tool support (MethylMix, ELMER, or both). This panel helps users quickly identify cancers showing strong hypermethylation that likely involves gene silencing, cancers showing strong hypomethylation that likely involves gene activation, and the degree of cross-tool consensus supporting methylation-driven dysregulation of the selected gene.
2.8.3 Cancer-Specific View: Methylation Distribution and Correlation
This view examines how methylation (β value) relates to gene expression within a selected cancer type. Selecting a cancer type updates all plots.
Components
A. Methylation–Expression Correlation Scatter Plot
This scatter plot displays β value (0–1) on the x-axis and TPM expression on the y-axis, with each point representing an individual sample color-coded by methylation status: red for hypermethylation, green for hypomethylation, blue for no methylation change, and grey for normal samples. Users can hover over points to view the β value, expression level, methylation category, and sample metadata. A negative correlation indicates that hypermethylation leads to reduced expression through gene silencing, while a positive correlation suggests that hypomethylation results in increased expression.
B. Expression by Methylation Status (Boxplot)
This boxplot shows TPM distribution across methylation categories (Hyper, Hypo, None), with hover details displaying full statistics including median, quartiles, fences, maximum, and minimum values. This visualization reveals whether expression levels differ significantly between methylation states, providing evidence for methylation-mediated transcriptional regulation.
C. β Value by Methylation Status (Boxplot)
This boxplot displays the β-value distribution for each methylation category, with hover functionality showing distribution details similar to the expression plots. This component confirms the expected separation between hyper-, hypo-, and unmethylated groups, validating the methylation classification used throughout the analysis.
D. Correlation Summary Panel
This panel displays the Spearman correlation coefficient (ρ) and associated p-value, measuring the strength and significance of the methylation–expression association within the selected cancer type.
β Value Reference
The β value is calculated as methylated probe intensity divided by the sum of methylated and unmethylated intensities, where 0 represents unmethylated state and 1 represents fully methylated state.

2.8.4 Methylation Summary Table
This table lists methylation statistics across cancer types, integrating results from MethylMix (gene-level) and ELMER (probe-level).
2.9 Gene Survival
2.9.1 Overview
The Gene Survival module evaluates how the selected gene influences patient prognosis across multiple cancer types. It provides two complementary analyses:- Single Gene Survival – assesses whether the gene alone is associated with patient outcomes across cancers.
- Survival Synergy Network – explores whether the gene shows cooperation (synergy) with other genes, where the combined expression affects survival more strongly than either gene alone.
Together, these tools help researchers identify genes with prognostic significance and uncover cooperative gene interactions relevant to cancer progression.
2.9.2 Single Gene Survival
This section analyzes how the expression level of the selected gene relates to patient survival across cancers.
User Inputs
- Survival Type: OS, PFI, DFI, DSS
- Stratification Method: median or mean expression cutoff

Survival Table
A table summarizes survival statistics across cancer types.Selecting a row generates Kaplan–Meier (KM) plots below. Use the table to quickly identify cancers where the selected gene shows significant prognostic impact.

Kaplan–Meier Plots
Selecting a cancer type displays two Kaplan–Meier plots showing the overall survival curve across all follow-up years and a 5-year survival curve, with the red curve representing the high expression group and the green curve representing the low expression group, where the y-axis shows survival probability and the x-axis shows time in months. The top of each plot displays the log-rank p-value and hazard ratio, and users can hover over the curves to view specific time points and survival probabilities at those moments. A hazard ratio greater than 1, where the red curve falls below the green curve, indicates that high expression is linked to poorer survival and suggests oncogene-like behavior, while a hazard ratio less than 1, where the red curve is above the green curve, indicates that high expression is linked to better survival and suggests tumor suppressor-like behavior.
2.9.3 Gene Survival Synergy Network
This section evaluates whether the selected gene shows synergistic survival effects with other genes, meaning their combined expression has a stronger impact on prognosis than either gene alone. A gene pair is considered synergistic when the combined hazard ratio (HR) of the two genes exceeds each individual gene's HR by a defined fold change (≥1.5× or ≥2×) and both genes have HRs in the same direction (both >1 or both <1), with synergy categories defined as positive synergy (HR > 1) where combined high expression is associated with worse prognosis, or negative synergy (HR < 1) where combined high expression is associated with better prognosis. Users can refine network results using filtering options including gene source (CGC, NCG, or all genes), synergistic effect level (≥1.5-fold or ≥2-fold), and HR direction (HR > 1 for risk-enhancing synergy, HR < 1 for protective synergy, or both). The network visualization displays nodes representing genes with the selected gene as the central node and edges representing synergistic survival relationships, where users can click a node to highlight connected synergistic partners or click on blank space to reset the view. This network identifies cooperative gene interactions that have stronger combined prognostic effects than individual gene behavior, revealing functional partnerships that may drive cancer progression or suppression through coordinated expression changes.
2.9.4 Synergistic Survival Comparison
This subsection provides survival curves for each synergistic gene pair identified in the network, examining how combined gene expression patterns affect patient outcomes. The table lists all gene pairs where the combined HR is greater than each single-gene HR with fold change greater than 1.0, though only pairs meeting the selected synergy threshold (≥1.5× or ≥2×) are considered true synergistic pairs in the network, and all p-values in this section are less than 0.05 to ensure statistical significance.
Filtering and Selection
Users can filter gene pairs by HR direction, choosing either HR > 1 where combined high expression predicts worse survival, or HR < 1 where combined high expression predicts better survival. The table displays the cancer type, Gene 1 and Gene 2 identifiers, HR fold change, log-rank p-values, and pairwise log2HR metrics for group comparisons, and users can select any row to generate the corresponding Kaplan–Meier plots.
Patient Grouping
Patients are stratified into four co-expression groups based on median expression of each gene: All.high (purple) where both genes are highly expressed, All.low (red) where both genes have low expression, High.low (blue) where the selected gene is high and the partner gene is low, and Low.high (green) where the selected gene is low and the partner gene is high. An additional 'Others' category combines All.low, High.low, and Low.high groups for two-group comparisons, allowing examination of how different co-expression patterns affect survival outcomes.
Kaplan–Meier Plots
Two complementary survival plots are displayed: the top plot shows a four-group comparison with separate survival curves for All.low, Low.high, High.low, and All.high groups, while the bottom plot presents a two-group comparison contrasting All.high versus all other groups combined. Both plots display hazard ratios (HR) and log-rank p-values, and users can hover over the curves to view specific time points and survival probabilities.
Interpretation
This analysis helps users determine whether the selected gene collaborates with specific partners to amplify poor prognosis through positive synergy or enhance favorable prognosis through negative synergy, separating mild combined effects from true synergistic behavior where gene pairs work cooperatively to influence patient outcomes beyond their individual contributions.

2.10 Gene miRNA
2.10.1 Overview
The Gene miRNA module visualizes and quantifies regulatory relationships between the selected gene and its associated miRNAs across multiple cancer types.
Interactions are compiled from 12 miRNA–target prediction tools and experimentally validated interactions recorded in miRTarBase, with flexible filtering to refine predicted vs. validated relationships.
- Gene–miRNA Interaction Network
- Gene–miRNA Correlation Table
2.10.2 Gene-miRNA Interaction Network
This interactive network displays predicted and experimentally validated regulatory interactions between the selected gene and miRNAs, integrating multiple data sources to provide comprehensive evidence for post-transcriptional regulation.
Data Sources
The network integrates three types of data: prediction tools (12 total) that identify interactions using multiple independent algorithms such as TargetScan, miRanda, RNAhybrid, DIANA-microT, and miRDB; validation data from miRTarBase containing experimentally confirmed interactions; and additional miRNA regulatory information from Chung et al. (Nucleic Acids Research, 2017) and YM500v3, which was used to identify negative correlations between driver genes and miRNAs in earlier DriverDB releases.
Network Representation
The network displays nodes and edges that represent genes, miRNAs, and their regulatory relationships. Nodes are color-coded with the selected gene shown in green and miRNAs predicted or validated to interact with the gene shown in yellow. All interactions are displayed as dotted lines. The type of interaction shown (validated, predicted, or both) depends on which checkboxes are enabled: experimentally validated interactions are sourced from miRTarBase, while predicted interactions are computationally derived with support based on the number of prediction tools that agree on the interaction.
Filtering Options
Users can refine the network using three filtering categories: gene source (CGC from Cancer Gene Census, NCG from Network of Cancer Genes, or all genes), minimum prediction support (≥6 tools, ≥8 tools, or ≥10 tools, where higher thresholds ensure stronger computational agreement), and a validation filter to show only validated interactions.
Interactions
The network provides interactive features including a search bar to enter gene or miRNA names and locate them within the network, node clicking to highlight only the selected node and its connected partners, and clicking empty space to reset to the full view.
Interpretation
This network allows users to identify miRNAs with high prediction support, identify miRNAs with experimental validation, explore potential regulatory repression mechanisms including interactions previously observed across cancers, and distinguish between predicted versus validated miRNA regulation of the selected gene. Together, the network provides a visual map of gene–miRNA interactions supported by both computational and experimental evidence, helping researchers understand post-transcriptional regulatory mechanisms that may influence gene expression in cancer.

2.10.3 Gene-miRNA Correlation Table
This table provides quantitative expression-based evidence supporting each predicted or validated interaction between the selected gene and miRNAs, displaying three correlation metrics that reflect gene–miRNA co-expression patterns across samples: Pearson correlation with p-value, Spearman correlation with p-value, and Kendall correlation with p-value. Negative correlations suggest miRNA-mediated repression of the gene, which is consistent with canonical miRNA targeting mechanisms where increased miRNA expression leads to decreased target gene expression, while the accompanying p-values indicate the statistical significance of the correlation strength and help users assess the reliability of each regulatory relationship.
2.11 Gene Protein
2.11.1 Overview
The Gene Protein module visualizes protein-level variation of the selected gene across cancers and examines how protein abundance relates to mRNA expression and post-translational modifications (PTMs).
Analyses are organized into three tabs:- Clinical Stages – grouped by clinical tumor stages
- Mutation Classes – grouped by mutation impact levels
- PTM Sites – grouped by specific phosphorylation sites (e.g., pY1068, pY1173)
All analyses support interactive exploration, including sample-level tooltips, togglable groups, and mRNA–protein scatter plots.
2.11.2 Clinical Stages
This tab evaluates how protein expression and mRNA–protein associations vary across clinical tumor stages.
Protein Expression by Clinical Stage (Pan-Cancer)
Purpose:Visualizes protein expression levels of the selected gene across all TCGA cancer types, grouped by stage.
Plot Features:- Boxplots display protein abundance for each cancer type (x-axis), grouped by stage (colors).
- Legend toggling: Show or hide specific stages (e.g., Stage I, Stage II, Stage III, Stage IV).
- Hover interactions:
- Hover over dots → sample-level details (sample ID, expression, tissue).
- Hover near box areas → summary statistics (median, Q1/Q3, upper/lower fences, min/max).
- Optional PTM selection: Choose to display None, pY1068, or pY1173 to inspect PTM-specific protein patterns.
Differences across stages may indicate stage-dependent dysregulation of protein abundance.

mRNA–Protein Correlation by Clinical Stage (Pan-Cancer)
Purpose:Assesses whether mRNA abundance explains protein expression patterns across cancers within each stage group.
Plot Features:- Bar chart showing Spearman correlation coefficients (ρ) between mRNA (FPKM-UQ) and protein expression across cancer types.
- Bars are grouped by clinical stage.
- Toggle individual stages via the legend.
- Hover interactions: Cancer type, stage, Spearman ρ, p-value.
- Click bar → opens a scatter plot (mRNA vs. protein), including the correlation and p-value.
- ρ > 0: mRNA and protein increase together → transcriptionally consistent regulation.
- ρ < 0: expression moves in opposite directions → post-transcriptional regulation or translational inhibition.

Cancer-Specific: Stage-Specific Protein Expression
This visualization shows protein expression patterns within a selected cancer type grouped by tumor stage, displaying a violin plot with stage groups (I–IV) on the x-axis and protein expression levels on the y-axis, where users can toggle stages using the legend and hover over dots to view sample-level information or hover over violin areas to see statistical summaries including median, quartiles, and fences. An accompanying statistical table compares pairs of stages with columns showing Group 1, Group 2, p-value, significance level, and sample counts, indicating whether stage-specific differences are statistically significant (p < 0.05) and helping users determine if protein expression changes progressively across disease stages or shows distinct patterns at specific stages of cancer development.
2.11.3 Mutation Classes
This tab evaluates how protein expression varies across mutation impact categories and how mutation classes influence mRNA–protein correlations.
Mutation impact groups:- High
- Moderate
- Low
- Modifier
- Normal tissue
- Tumors without mutation
Protein Expression by Mutation Class (Pan-Cancer)
Purpose:Visualizes protein expression across cancers grouped by mutation impact level.
Plot Features:- Boxplots grouped by impact class,, one set per cancer type
- Legend toggling for impact classes
- Hover for sample-level details and boxplot summary statistics
- Optional PTM filtering (None, pY1068, pY1173)
Allows users to assess whether specific impact classes (e.g., high-impact mutations) correspond to altered protein levels.

mRNA–Protein Correlation by Mutation Class (Pan-Cancer)
Purpose:Examines mRNA–protein concordance across mutation-defined sample groups.
Features:- Bar chart of Spearman ρ for each cancer type, grouped by mutation impact
- Hover for cancer type, impact class, ρ, p-value
- Click bar → opens the corresponding mRNA–protein scatter plot
- Positive ρ: protein expression tracks mRNA → transcriptionally driven response
- Negative ρ: mutation-class–specific post-transcriptional or PTM-dependent regulation

Cancer-Specific: Mutation-Impact–Specific Protein Expression
This visualization shows protein expression differences within a selected cancer type grouped by mutation class, displaying a violin plot with mutation impact class on the x-axis and protein expression levels on the y-axis, where users can hover for statistical summaries and sample information and toggle impact classes using the legend. An accompanying statistical table provides pairwise comparisons between impact classes with columns showing Group 1, Group 2, p-value, significance level, and sample counts. This analysis helps identify whether high-impact mutation carriers show altered protein levels relative to other mutation groups, revealing whether mutations influence not only gene expression at the transcript level but also at the protein level, which may have more direct functional consequences for cancer phenotypes.
2.11.4 PTM Sites
This tab evaluates how post-translational modifications (PTMs)—specifically phosphorylation sites—modify the relationship between mRNA and protein expression.
mRNA-Protein Correlation by PTM Site (Pan-Cancer)
Purpose:Assesses how PTMs (e.g., pY1068, pY1173) influence mRNA–protein coupling across cancers.
Plot Features:- Bar chart of Spearman correlation coefficients (ρ) across cancer types
- Groups correspond to PTM sites:
- None (total protein)
- pY1068
- pY1173
- Toggle PTM sites using the legend
- Hover for Cancer type, PTM site, ρ, p-value
- Click bar → opens a PTM-specific mRNA–protein scatter plot
- Positive ρ (> 0):PTM-site–specific protein levels track mRNA → transcriptionally driven regulation
- Negative ρ (< 0):Protein/PTM levels diverge from mRNA → post-transcriptional or PTM-dependent modulation
(e.g., phosphorylation buffering, kinase pathway activation independent of transcript levels)

2.12 Gene Multi-omics
2.12.1 Overview
The Gene Multi-Omics interface summarizes how multiple molecular layers—including gene expression, mutation, CNV, methylation, protein, and miRNA—converge to support the user-selected gene as a potential driver across cancers.
All results reflect outputs from multi-omics integration tools, providing a cross-layer view of biological evidence.
- Integrated Multi-Omics Overview
- Omics Connectivity Network
- List of Multi-Omics Driver Events
Each section offers complementary insights into how strongly the selected gene is supported as a multi-omics driver.
2.12.2 Integrated Multi-Omics Overview
This section aggregates driver evidence from multi-omics prediction tools and displays where (cancer types) and how (omic layers) the selected gene is supported, with visualization presented through three coordinated components that together provide a comprehensive view of multi-layered evidence.
Top-Right Bar Chart — Tool Support Across Cancers
This bar chart shows how many integration tools identified the selected gene as a driver in each cancer type, with users able to hover to view the cancer type and tool support count. Taller bars indicate stronger cross-tool agreement within that particular cancer, suggesting the gene is consistently recognized as a driver across multiple computational methods in those cancer types.
Bottom-Left Bar Chart — Tool Support Across Omics Layers
This bar chart summarizes the number of supporting tools for each omic type, including mutation, CNV, methylation, expression, and miRNA, with hover functionality displaying the omics type and tool count. This component highlights whether the gene is supported across multiple molecular mechanisms, revealing the diversity of dysregulation patterns affecting the gene.
Bottom-Right Combination Matrix — Cross-Omics Evidence
This matrix displays cancer types as rows and omics layers as columns, where each solid dot indicates that the gene is identified as significant in the corresponding omic type for that cancer dataset, with filled dots representing support by at least one integrative tool. Users can hover over dots to view cancer type, omics type, and evidence details. Dense dot patterns indicate broad multi-omics support across many cancer types, wide spread across omics columns suggests convergent dysregulation of the gene through multiple molecular mechanisms, and dense rows highlight specific cancers where the gene plays a multi-omics functional role with alterations spanning multiple biological layers.

2.12.3 Omics Connectivity Network
This network visualizes the associations linking the selected gene, relevant omics layers, and supported cancer types through an interactive graph representation. Nodes in the network include the central gene node, omics category nodes, and cancer type nodes, while edges represent relationships identified by integration tools such as mutation-driven amplification, methylation-associated silencing, and CNV-driven expression shifts. Users can click on a node to highlight connected omics and cancer relationships or hover over nodes to view the node type and related connections. Nodes with many edges represent key integration points, indicating that the gene influences multiple omics layers or participates in multi-cancer dysregulation, revealing the breadth and complexity of the gene's role across different molecular mechanisms and cancer contexts.
2.12.4 Multi-Omics Driver Event Table
This detailed table lists all driver events supported by multi-omics tools for the selected gene, providing comprehensive information about the molecular context and reproducibility of each driver event. High nTool values indicate strong cross-tool reproducibility where multiple computational methods independently identify the same driver event, while CGC/NCG = Yes indicates independent biological support of cancer relevance through inclusion in curated cancer gene databases. The presence of multiple omics layers for a gene demonstrates multi-layer dysfunction characteristic of driver genes, where alterations span genomic, epigenomic, and transcriptomic levels. This table allows users to quickly identify the specific omics drivers and cancer contexts in which the gene is strongly supported, enabling prioritization of the most robust and biologically relevant driver events for further investigation or therapeutic targeting.
3. Customized Analysis
3.1 Overview
The Customized Analysis module enables researchers to perform user-defined analyses using clinical subgroups, gene features, and survival outcomes.
Unlike the Cancer and Gene modules—which summarize fixed results—Customized Analysis allows flexible, interactive, and hypothesis-driven exploration.
- Subgroup Comparison Analyses
- Survival Analyses
- Multi-Omics Driver Analysis
- Prognostic Signature Identification
- Multivariate Survival Analysis
Below is detailed help for the Subgroup Comparison Analyses section.
3.2 Subgroup Comparison Analyses
Subgroup Comparison Analyses evaluate how gene-level molecular features differ across clinically defined patient subgroups.
Users define subgroups using any combination of clinical parameters (e.g., stage, grade, receptor status), and analyses are performed per selected gene and dataset.
- Expression
- Mutation
- CNV
- Methylation
Each analysis helps uncover biology associated with disease progression, risk groups, treatment response, or other clinically important factors.
3.2.1 Expression Subgroup Comparison
This analysis assesses whether gene expression varies across clinically defined patient subgroups within a cancer dataset.
Workflow
- Select a gene of interest.
- Choose a dataset.
- Define the analysis cohort by selecting one or more clinical criteria (e.g., Stage I+II, ER−, Grade 3).
- Users may select multiple criteria simultaneously; sample counts update automatically.
- Choose a subgroup factor (e.g., stage, grade, receptor status), which determines the x-axis grouping in plots.
Output: Expression Comparison Across Clinical Subgroups
Violin Plot (log₁₀ TPM)- Displays expression distributions after log transformation.
- Log scale reduces extreme variance and highlights differences between subgroup distributions.
- Hover for summary statistics (median, quartiles, fences) or sample-level data.
- Shows raw expression values without transformation.
- Useful for interpreting absolute expression magnitude.
- Hover to view sample-level details.
Pairwise comparisons are automatically generated across subgroup levels.
Columns include:
- Group1 / Group2
- p-value
- Significance
- ns (≥0.05)
- * (<0.05)
- ** (<0.01)
- *** (<0.001)
- **** (<0.0001, optional)
- Sample counts
Interpretation
Together, the violin plots and comparison table help determine whether gene expression differs meaningfully across clinical categories such as:- Tumor stage
- Histologic grade
- Receptor/HER2 status
- Molecular subtype
- Treatment response groups

3.2.2 Mutation Subgroup Comparison
This analysis evaluates whether mutation frequency of the selected gene differs between two clinically defined patient groups.
Workflow
- Select a gene and dataset.
- Define Group 1 and Group 2 using one or more clinical criteria (e.g., Stage I vs Stage III–IV, ER+ vs ER−).
- Each group must contain ≥20 samples to ensure statistical validity.
- Run the analysis to generate contingency and statistical results.
Output
Mutation Contingency TableDisplays mutation counts for each group:
- Group 1 Mutated / Wild-type
- Group 2 Mutated / Wild-type
- Total sample counts
This table summarizes how mutation events are distributed across the two subpopulations.
Fisher’s Exact Test StatisticsComputed to determine whether mutation frequencies differ significantly.
Outputs include:
- Odds Ratio (OR)
- OR > 1 → mutations more common in Group 1
- OR < 1 → mutations more common in Group 2
- 95% Confidence Interval (CI)
- p-value (Fisher’s Exact Test)
Use this analysis to determine whether clinical subgroups differ in mutation burden for the selected gene.
Biological questions supported include:
- Are late-stage tumors more mutated?
- Do ER− patients have higher mutation frequency?
- Are responders and non-responders genetically distinct?

3.2.3 CNV Subgroup Comparison
The CNV Subgroup Comparison evaluates whether copy number variation (CNV) patterns differ between two clinically defined patient groups.
Events are categorized as Gain, Loss, or None (neutral).
Workflow
- Select a gene and dataset.
- Define Group 1 and Group 2 using one or more clinical criteria (e.g., Stage I vs Stage III–IV).
- Each group must include ≥20 samples.
- Run the analysis to generate CNV contingency tables and Fisher’s Exact Test results.
Output
CNV Contingency Tables (Four Comparisons)- All CNV Categories Combined
- Shows counts of Gain, Loss, and None in both groups.
- Provides an overall view of CNV distribution.
- Gain vs. Loss
- Excludes neutral samples.
- Tests whether amplification vs. deletion trends differ.
- Gain vs. None
- Compares Gain against neutral CNV states.
- Loss vs. None
- Compares Loss against neutral states.
- Odds Ratio (OR):
- OR > 1 → CNV event more common in Group 1
- OR < 1 → CNV event more common in Group 2
- 95% CI
- p-value (Fisher’s Exact Test)
Interpretation
This analysis reveals whether the selected gene exhibits different CNV profiles across clinical subpopulations—for example:- Amplification enriched in late-stage cancers
- Deletion enriched in a specific molecular subtype
- Neutral copy number predominance in certain risk groups

3.2.4 Methylation Subgroup Comparison
The Methylation Subgroup Comparison evaluates whether DNA methylation levels (β-values) differ across clinically defined patient subgroups.
Workflow
- Choose a gene and dataset.
- Filter the cohort by selecting clinical criteria.
- Select a subgroup factor (e.g., stage, grade, receptor status).
- This determines the x-axis grouping.
- Run the analysis to generate violin plots and statistical comparisons.
Output
Methylation Violin Plot (β-values)- Y-axis: β-value (0–1)
- 0 = unmethylated
- 1 = fully methylated
- X-axis: subgroup factor categories
- Each violin shows the methylation distribution within each subgroup.
- Hover for summary statistics (median, quartiles, fences) or sample-level details.
Includes pairwise subgroup comparisons:
- Group 1 / Group 2
- p-value
- Significance (ns, *, **, ***, ****)
- Sample counts per subgroup
Interpretation
This analysis helps determine whether epigenetic regulation of the gene differs across patient groups—for example:- Hypermethylation enriched in high-grade tumors
- Hypomethylation associated with specific receptor status
- Stage-dependent methylation differences

3.3 Survival Analyses
Survival Analyses evaluate how mutations, gene expression levels, or miRNA expression levels influence patient outcomes within a user-defined subpopulation. Users define the patient cohort via clinical criteria and select stratification methods. This section begins with Mutation-Based Survival Analysis, followed by Expression-Based Survival Analysis and miRNA-Based Survival Analysis.
3.3.1 Mutation-Based Survival Analysis
The Mutation-Based Survival Analysis assesses whether mutations in the selected gene list are associated with survival differences in a clinically defined subpopulation.
Workflow
- Input a gene list.
- Select a dataset.
- Use clinical criteria to filter the patient cohort (e.g., Stage II only, ER+ only).
- This determines which patients will be evaluated.
- The results tab then allows you to dynamically select:
- Stratification method (Mutation vs. Wild type or Number of mutated genes)
- Time interval (All follow-up or 5-year)
These two options control both the survival table and the Kaplan–Meier plots below.
Output
A. Mutation Oncoprint
This visualization provides a visual summary of mutation patterns across patients, with rows representing genes from the input list, columns representing individual patients, and cells color-coded by mutation impact where red indicates high impact, blue indicates moderate impact, and additional colors are used as applicable. Side panels provide complementary information: the left panel displays the percentage of mutated samples for each gene, while the top panel shows mutation burden or impact summary, often displayed as a combination impact score (e.g., 0–2). This visualization quickly shows which genes are frequently mutated and how mutation profiles vary across patients within the selected cohort.
B. Survival Control Panel
Located directly above the survival table and Kaplan–Meier plots, this panel includes dropdown menus for selecting the stratification method (Mutation vs. Wild type or By number of mutated genes) and time interval (All follow-up or 5-year survival). Changing these settings immediately updates the KM curves to reflect the selected analysis parameters.
C. Survival Statistics Table
This comprehensive table summarizes survival analysis results for each cancer type, gene or gene set, and survival endpoint combination. Key information includes the cancer type abbreviation, the gene(s) evaluated under the selected stratification, the outcome analyzed (OS for Overall Survival, PFI for Progression-Free Interval, DFI for Disease-Free Interval, DSS for Disease-Specific Survival), the stratification method used, which groups serve as the comparison factor versus reference, log-rank and Cox p-values for both all follow-up time and the 5-year interval, hazard ratios and their log2 transforms for both time periods, and sample counts for mutated and wild-type groups. HR values greater than 1 indicate the mutated group has worse prognosis, HR values less than 1 indicate the mutated group has better prognosis, and p-values less than 0.05 indicate significant survival differences between groups.
D. Kaplan–Meier (KM) Survival Plots
For every analysis, four Kaplan–Meier plots are generated—one for each survival endpoint: OS (Overall Survival), PFI (Progression-Free Interval), DFI (Disease-Free Interval), and DSS (Disease-Specific Survival). The KM curves automatically update based on the selected stratification method (Mutation vs. Wild type or Number of mutated genes) and time interval (All follow-up or 5-year survival). Each KM plot includes color-coded survival curves for the selected groups, survival probability over time in months, log-rank test p-value, and hover interaction to view timepoint-specific survival values. These four KM plots allow users to visually compare survival differences across mutation-defined groups for all major survival outcomes, with the multi-endpoint output being particularly useful for identifying consistent trends or endpoint-specific associations across different measures of patient prognosis.

3.3.2 Expression-based Survival Analysis
The Expression-Based Survival Analysis evaluates whether gene expression levels are associated with survival outcomes (OS, PFI, DSS, DFI) in a user-defined patient subpopulation, allowing users to stratify patients based on gene expression and examine survival differences across clinical groups.
Workflow
- Select gene(s)
Enter one or multiple genes whose expression will be used for group stratification. - Select dataset
Choose the cancer dataset on which survival analysis will be performed. - Define patient subpopulation
Use clinical criteria (e.g., stage, grade, ER/PR/HER2 status, molecular subtype) to filter the cohort.
These filters determine which patients will be included in the analysis. - Submit
The system loads the analysis interface.
Output
A. Stratification Control Panel
B. Survival Table
This table summarizes survival statistics for each gene, cancer type, and survival endpoint. Key metrics include hazard ratios (HR) and p-values for both all follow-up and 5-year intervals, expression cutoff thresholds, stratification methods, and sample counts for high and low-expression groups. HR > 1 indicates high expression is associated with worse prognosis, HR < 1 indicates better prognosis, and p < 0.05 indicates significant survival differences.
C. Kaplan-Meier (KM) Survival Plots
For each survival outcome, KM curves visualize survival differences between expression-defined groups, with one curve per group (High vs Low, All-high vs Others, etc.), log-rank p-value and HR displayed on the plot, and curves automatically updating based on cutoff method, grouping method, and time interval (5-year vs all follow-up). These plots show whether expression differences translate into clinically meaningful survival divergence and help users assess the prognostic value of the selected gene(s) in the filtered patient cohort.
D. Boxplots of Gene Expression
These boxplots summarize the expression distributions of the selected gene(s) within the filtered cohort, displaying TPM or log10(TPM) values with hover functionality to view sample-level values and summary statistics including median, Q1, Q3, fences, minimum, and maximum. This visualization helps confirm that expression-defined patient groups are meaningfully different in terms of gene expression levels before analyzing survival outcomes, ensuring that stratification produces biologically distinct groups for comparison.

3.3.3 miRNA-based Survival Analysis
Workflow
- Input a miRNA (e.g., hsa-miR-21-5p) to define the expression feature.
- Select an analysis framework: Cox Uni, Cox Multi, or Cure Model.
- After choosing the framework, configure the analysis using dropdown menus:
- Cancer type – choose the cohort to analyze
- Survival endpoint – select the outcome type
- Survival time – follow-up time scale on the x-axis (months)
- Stratification method – split patients into groups based on miRNA expression (e.g., High vs Low)
- View results as survival/hazard curves and (when applicable) risk estimates.
Output
A. Cox Uni Results (Univariate Cox Analysis)This analysis displays two complementary visualizations when Cox Uni is selected and dropdown menus are configured. The Kaplan–Meier (KM) survival plot shows survival probability (y-axis) over time in months (x-axis) for miRNA-defined groups (e.g., High vs Low), allowing users to compare curve separation between groups to assess outcome differences and use the reported log-rank p-value to evaluate whether the group difference is statistically supported. The cumulative hazard plot shows cumulative hazard (accumulated risk of the event) over months for the same stratified groups, where steeper curves indicate risk accumulating more quickly and can be used alongside the KM plot to view group differences in terms of risk accumulation rather than survival probability. Clear separation between High versus Low groups suggests the miRNA is associated with prognosis without clinical adjustment, indicating a univariate association between miRNA expression and patient outcomes.
B. Cox Multi (Multivariate Cox Analysis with Clinical Adjustment)This analysis displays two complementary visualizations when Cox Multi is selected and dropdown menus are configured. The adjusted survival curve shows model-predicted survival probability (y-axis) over months (x-axis) for miRNA-defined groups after adjusting for available clinical covariates in that cohort, allowing users to assess whether group separation persists after clinical adjustment and provides evidence that the miRNA offers prognostic information beyond standard clinical variables within the available covariates. The forest plot displays hazard ratios (HRs) with 95% confidence intervals for the miRNA group term (e.g., High vs Low) and each included clinical covariate that was automatically selected based on cohort availability, where HR > 1 indicates higher risk (worse outcome), HR < 1 indicates lower risk (better outcome), a dashed vertical line at HR = 1 indicates no effect, and wider confidence intervals indicate greater uncertainty. A significant miRNA-group HR after adjustment suggests the miRNA is an independent prognostic factor given the included covariates, providing evidence that miRNA expression contributes prognostic value beyond traditional clinical variables.
C. Cure ModelThis analysis displays a cure-model survival curve when Cure Model is selected and dropdown menus are configured, showing cure-model–estimated survival probability (y-axis) over months (x-axis) for miRNA-defined groups. Users can compare curves to evaluate group-specific outcome differences under a model designed to capture long-term survival patterns and look for late-time plateaus that can reflect sustained survival behavior. Group separation indicates prognostic differences in a framework designed for long-term survival dynamics, which can be particularly informative when standard proportional hazards assumptions may not fully reflect the data and when a subset of patients may experience extended disease-free survival.
3.4 Multi-omics Driver Analysis
Overview
The Multi-omics Driver Analysis identifies driver events that differ between two clinically defined patient groups, integrating:- Gene expression
- Mutation
- Copy number variation (CNV)
- Methylation
It highlights which genes and pathways are most likely driving group differences (e.g., responders vs non-responders, early vs late stage) and how consistently they are supported across omics types and tools.
Use this analysis when you want to understand which genomic and epigenomic alterations underlie clinical subgroup differences.
Workflow
- Select dataset
Choose the cancer dataset to analyze. - Define Group 1 and Group 2
Use clinical criteria (e.g., stage, grade, response status, receptor subtype) to define two patient groups.- Each group should contain ≥ 20 samples for robust statistical analysis.
- Choose gene set option (Gene Dataset)
- All – analyze all eligible genes in the dataset
- CGC – restrict to genes listed in Cancer Gene Census (CGC)
- NCG – restrict to genes listed in Network of Cancer Genes (NCG 6.0)
- Run analysis
The results page will display driver events and multiple integrated visualizations.
Output
A. Multi-layer Driver-Function Relationship Diagram & Driver Summary TableMulti-layer Driver-Function Relationship Diagram
This network-like diagram connects the selected cancer dataset, omics layers (mRNA, Mutation, CNV, Methylation), driver genes, and functional/pathway terms such as GO terms to illustrate the relationships between molecular alterations and biological functions. The structure flows hierarchically: the cancer node connects to each omics node (mRNA, Mutation, CNV, Methylation), each omics node connects to driver genes identified in that layer, and driver genes connect to GO term or function nodes representing enriched pathways or processes. Users can trace paths from clinical groups through omics alterations to driver genes and finally to biological functions, identifying which omics layers contribute most to observed group differences, which driver genes are shared across omics layers, and which biological functions and pathways are most impacted by these alterations.
Driver Summary TableThis table lists all identified drivers with their cancer type/dataset, omics layer (mRNA, Mutation, CNV, Methylation), driver gene symbol, Cancer Gene Census (CGC) status, Network of Cancer Genes (NCG) status, integration method that detected the driver, number of tools supporting this driver event (nTools), and associated Gene Ontology terms indicating pathways or functions. Higher nTools values indicate stronger cross-tool evidence for a driver event, CGC/NCG = Yes provides additional external support as a known cancer gene, and GO_term entries reveal potential biological roles and affected pathways. Together, the diagram and table summarize how multi-omics drivers are identified and what functional roles they may play in cancer biology.

B. Distribution of Drivers Across Omics Types and Tools
This section helps evaluate how strongly each driver is supported across omics layers and computational methods through two complementary visualizations.
Omics-by-Gene Tool Support Heatmap (Left)This heatmap displays genes as rows and omics types (mRNA, Mutation, CNV, Methylation) as columns, with each cell value representing the number of tools that identified that gene as a driver in that specific omic layer. Users can hover on cells to see the exact tool count for each gene-omic combination, enabling identification of robust multi-omics drivers including genes with high support across multiple omics layers and genes supported by many tools in at least one omic category. This visualization helps prioritize genes based on the breadth and depth of computational evidence supporting their driver status.
Tools per Gene by Omics Bar Plot (Right)This bar plot displays each driver gene with bar height representing the total number of tools that support that gene as a driver, with color coding indicating the contributions from different omics layers (mRNA, Mutation, CNV, Methylation). Users can compare tool support between genes to identify the most robustly detected drivers and use the legend to toggle specific omics types on or off, allowing focused examination of particular molecular layers. Genes with high tool support across several omics types are high-confidence multi-omics drivers, as convergent evidence from multiple computational methods and molecular mechanisms strengthens the reliability of their identification as functionally important cancer genes.

C. Coverage and Consistency of Multi-omics Identification Tools
This section focuses on tools rather than genes, evaluating how comprehensively tools cover omics layers and how consistent their driver identifications are across methods.
Omics-by-Tool Coverage Heatmap (Left)This heatmap displays tools as rows and omics types as columns, with each cell showing the proportion of drivers in each omic layer detected by each specific tool. Users can examine which tools have broad coverage across multiple omics types versus those with more selective detection patterns focused on particular molecular layers, and identify tools that contribute most substantially to driver identification within a specific omics type. This visualization reveals the complementary nature of different computational approaches and helps users understand which tools are most effective for detecting drivers in each molecular context.
Tool Overlap Distribution Plot (Right)This bar chart displays the number of tools (x-axis) versus the number of genes detected by that many tools (y-axis), revealing the degree of consensus among computational methods in driver identification. Genes detected by multiple tools are generally more reliable as convergent evidence from independent methods strengthens confidence in their driver status, while a right-shifted distribution with more genes supported by many tools suggests strong cross-tool consistency in the analytical pipeline. This plot helps users assess the overall reproducibility of driver detection and identify which genes have the most robust computational support across the integrated multi-omics framework.

3.5 Prognostic Signature Identification
3.5.1 Overview
The Prognostic Signature Identification analysis constructs a survival-predictive gene signature from a user-provided gene list. Using LASSO (Least Absolute Shrinkage and Selection Operator) and Random Forest models, the analysis identifies survival-associated genes, builds a multigene risk-score model, and evaluates its predictive performance through survival statistics, ROC curves, risk stratification plots, and feature-selection diagnostics.
Use this analysis if you already have a candidate gene list and want to determine:- Which genes are most predictive of survival
- How these genes can be combined into a prognostic signature
- How well the signature stratifies patients into risk groups
- What biological functions are enriched among signature genes
3.5.2 Workflow
1. Input a Gene List
Users may provide a list of candidate genes using either method:- Type or paste genes directly into the text box
- Upload a .txt file containing one gene symbol per line
These genes will be used to identify survival-related markers and construct the prognostic signature.
2. Select Dataset Settings
After submitting the gene list, users configure all analysis settings on the results page.
Select a TissueChoose a broad tissue category (e.g., Breast, Lung, Colon).
This filters the available cancer datasets.
Select Cancer Type
From the filtered list, select a TCGA (or other) cancer dataset for training the prognostic signature model.
3. Select Data Type(s) for Model Construction
Users may choose one or multiple omics types used for signature construction:- RNA expression
- Copy Number Variation (CNV)
- Mutation
- Methylation
Selected data types define which molecular features contribute to the LASSO and Random Forest survival models.
4. Select a Survival Endpoint
Choose the patient outcome to be modeled:- Overall Survival (OS)
- Progression-Free Interval (PFI)
- Disease-Free Interval (DFI)
- Disease-Specific Survival (DSS)
The endpoint determines how prognostic performance is evaluated.
5. Define Patient Subpopulation (Clinical Criteria Filter)
Filter the dataset to analyze a specific patient subpopulation by applying one or more clinical criteria.
Each criterion includes multiple groups with sample counts.
Users may combine multiple criteria to define a precise analysis cohort.
3.5.3 Results Overview
- Statistical Summary Table
- Kaplan-Meier Plot
- Time-Dependent ROC Curves
- Prognostic Risk Score Model
- Risk Heatmap
- Lambda Screening Plot
- Functional Annotation Barplots
- Shrinkage Gene List
3.6 Multivariate Survival Analysis
3.6.1 Overview
In the Multivariate Survival Analysis, over one hundred clinical factors are available for selection to construct a comprehensive prognostic model. If you are interested in comparing specific candidate gene(s) with well-known clinical prognostic biomarkers, this analysis will construct a multivariate model with customized clinical factors in the CoxPH framework. The generated report includes corresponding statistical results, Kaplan–Meier plots, and point-estimated values of all factors displayed in a forest plot.
3.6.2 Workflow
1. Input a Gene List or Signature list
Users choose the input type—either gene name or signature—and provide their list of candidate genes by typing or pasting gene symbols directly into the text box. These genes will be used to identify survival-related markers and construct the prognostic signature for the selected cancer cohort.
2. Select Dataset Settings
After submitting the gene list, users configure all analysis settings on the results page.
Select a TissueChoose a broad tissue category (e.g., Breast, Lung, Colon).
This filters the available cancer datasets.
From the filtered list, select a TCGA (or other) cancer dataset for training the prognostic signature model.
3. Select Confounding Factors
This section determines how clinical variables are handled in the survival model.Users:
- Select clinical factors (e.g., Age, Stage, Gender, Grade) to adjust for.
- Optionally enable: “Confounding factors selected by LASSO”
- Unchecked:
All selected confounding factors are forced into the Cox model—always included. - Checked:
Selected confounders are also subjected to LASSO.
LASSO selects only the clinical factors that meaningfully contribute to prognosis, shrinking others to zero.
- Full adjustment (all selected confounders included)
- Sparse adjustment (LASSO optimizes both genes and clinical variables)
4. Select Data Type(s) for Model Construction
Users may choose one or multiple omics types used for signature construction:- RNA expression
- Copy Number Variation (CNV)
- Mutation
- Methylation
Selected data types define which molecular features contribute to the LASSO and Random Forest survival models.
5. Select a Survival Endpoint
Choose the patient outcome to be modeled:- Overall Survival (OS)
- Progression-Free Interval (PFI)
- Disease-Free Interval (DFI)
- Disease-Specific Survival (DSS)
The endpoint determines how prognostic performance is evaluated.
6. Define Patient Subpopulation (Clinical Criteria Filter)
Filter the dataset to analyze a specific patient subpopulation by applying one or more clinical criteria.
Each criterion includes multiple groups with sample counts.
Users may combine multiple criteria to define a precise analysis cohort.
3.6.3 Results Overview
The analysis generates comprehensive results for each selected data type, including a summary table with statistical metrics, Kaplan–Meier plots showing survival curves for risk-stratified groups, and forest plots displaying hazard ratios with confidence intervals for all genes and clinical factors included in the final multivariate model.
Download
DriverDBv4 provides four summaries of driver genes for users to download: mutation drivers defined by 9 mutation tools in various cancers, CNV drivers defined by 2 tools in various cancers, methylation drivers defined by 2 tools in various cancers, and multi-omics drivers defined by 8 tools.
Dataset
| Project Name | Cancer Name | Tissue |
| ACC-TCGA | Adrenocortical_carcinoma | Adrenal_gland |
| BLCA-CN-ICGC | Bladder_urothelial_carcinoma | Bladder |
| BLCA-TCGA | Bladder_urothelial_carcinoma | Bladder |
| BOCA-FR-ICGC | Ewing_sarcoma | Bone |
| BOCA-UK-ICGC | Osteosarcoma_chondrosarcoma_rare_subtypes | Bone |
| BPLL-FR-ICGC | B_Cell_Prolymphocytic_Leukemia | Blood |
| BRCA-EU-ICGC | Breast_carcinoma | Breast |
| BRCA-FR-ICGC | Breast_carcinoma | Breast |
| BRCA-KR-ICGC | Breast_carcinoma | Breast |
| BRCA-SCANB | Breast_carcinoma | Breast |
| BRCA-TCGA | Breast_carcinoma | Breast |
| BRCA-UK-ICGC | Breast_carcinoma | Breast |
| BTCA-JP-ICGC | Biliary_Tract_cancer | Gall bladder |
| BTCA-SG-ICGC | Biliary_Tract_cancer | Gall bladder |
| CESC-TCGA | Cervical_squamous_cell_carcinoma | Cervix |
| CHOL-TCGA | Cholangiocarcinoma | Bile_duct |
| LCLL-ES-ICGC | Chronic_Lymphocytic_Leukemia | Blood |
| CMDI-UK-ICGC | Chronic_myeloid_disorders | Blood |
| COAD-TCGA | Colon_adenocarcinoma | Colon |
| COCA-CN-ICGC | Colorectal_cancer | Colon |
| DLBC-TCGA | Lymphoid_neoplasm_diffuse_large_b_cell_lymphoma | Blood |
| PRAD-DE-ICGC | Prostate_adenocarcinoma | Prostate |
| ESCA-CN-ICGC | Esophageal_carcinoma_squamous | Esophagus |
| LIHC-CN-ICGC | Hepatocellular_carcinoma_HBV_associated | Liver |
| ESCA-TCGA | Esophageal_carcinoma | Esophagus |
| ESCA-UK-ICGC | Esophageal_carcinoma_adenocarcinoma | Esophagus |
| GACA-CN-ICGC | Gastric_cancer | Stomach |
| GACA-JP-ICGC | Gastric_cancer | Stomach |
| GBM-TCGA | Glioblastoma_multiforme | Brain |
| HNSC-TCGA | Head_and_neck_squamous_cell_carcinoma | Head_and_neck |
| KICH-TCGA | Kidney_chromophobe | Kidney |
| KIRC-TARGET | kidney_clear_cell_sarcoma | Kidney |
| KIRC-TCGA | Kidney_renal_clear_cell_carcinoma | Kidney |
| KIRP-TCGA | Kidney_renal_papillary_cell_carcinoma | Kidney |
| LALL-TARGET | Acute_lymphoblastic_leukemia | Blood |
| LAML-CN-ICGC | Acute_myeloid_leukemia | Blood |
| LAML-KR-ICGC | Acute_myeloid_leukemia | Blood |
| LAML-TARGET | Acute_myeloid_leukemia | Blood |
| LAML-TCGA | Acute_myeloid_leukemia | Blood |
| LGG-TCGA | Low_grade_glioma | Brain |
| LIHC-FR-ICGC | Hepatocellula_adenoma | Liver |
| LIHC-FR-ICGC | Hepatocellular_carcinoma_Secondary_to_alcohol_and_adiposity | Liver |
| LIHC-TCGA | Liver_hepatocellular_carcinoma | Liver |
| LIHC-FR-ICGC | Hepatocellular_carcinoma_Secondary_to_alcohol_and_adiposity | Liver |
| LIHC-JP-ICGC | Hepatocellular_carcinoma_Virus_associated | Liver |
| LIHC-JP-ICGC | Hepatocellular_carcinoma_Virus_associated | Liver |
| SARC-FR-ICGC | Leiomyosarcoma | Soft_tissue |
| LUAD-TCGA | Lung_adenocarcinoma | Lung |
| LUSC-CN-ICGC | Lung_squamous_cell_carcinoma | Lung |
| LU-KR-ICGC | Adenocarcinoma_Squamous_cell_carcinoma | Lung |
| LUSC-TCGA | Lung_squamous_cell_carcinoma | Lung |
| MALY-DE-ICGC | Germinal_center_B_cell_derived_lymphomas | Blood |
| SKCM-AU-ICGC | Skin_cutaneous_melanoma | Skin |
| MESO-TCGA | Mesothelioma | Mesothelium |
| HNSC-CN-ICGC | Nasopharyngeal_carcinoma | Nasopharynx |
| NBL-TARGET | Neuroblastoma | Nervous_system |
| NKTL-SG-ICGC | T_cell_and_NK_cell_lymphoma | Blood |
| HNSC-IN-ICGC | Oral_cancer | Head_and_neck |
| BOCA-TARGET | Osteosarcoma | Bone |
| OV-AU-ICGC | Ovarian_cancer | Ovary |
| OV-TCGA | Ovarian_serous_cystadenocarcinoma | Ovary |
| PAAD-TCGA | Pancreatic_adenocarcinoma | Pancreas |
| PAAD-AU-ICGC | Prostate_ductal_adenocarcinoma | Pancreas |
| PAAD-CA-ICGC | Prostate_ductal_adenocarcinoma | Pancreas |
| PAEN-AU-ICGC | Endocrine_neoplasms | Pancreas |
| PAEN-IT-ICGC | Endocrine_neoplasms | Pancreas |
| PBCA-DE-ICGC | Pediatric_Medulloblastoma_and_Pilocytic_Astrocytoma | Brain |
| PBCA-US-ICGC | Pediatric_Brain_Tumor | Brain |
| PCPG-TCGA | Pheochromocytoma_and_paraganglioma | Adrenal_gland |
| PEME-CA-ICGC | Pediatric_Medulloblastoma | Brain |
| PRAD-CA-ICGC | Prostate_adenocarcinoma | Prostate |
| PRAD-CN-ICGC | Prostate_adenocarcinoma | Prostate |
| PRAD-FR-ICGC | Prostate_adenocarcinoma | Prostate |
| PRAD-TCGA | Prostate_adenocarcinoma | Prostate |
| PRAD-UK-ICGC | Prostate_adenocarcinoma | Prostate |
| READ-TCGA | Rectum_adenocarcinoma | Rectum |
| KIRC-CN-ICGC | Kidney_renal_clear_cell_carcinoma | Kidney |
| RECA-EU-ICGC | Renal cell carcinoma (Focus on but not limited to clear cell subtype) | Kidney |
| RT-TARGET | Rhabdoid_tumors | Kidney |
| SARC-TCGA | Sarcoma | Soft_tissue |
| SKCM-BR-ICGC | Skin_cutaneous_melanoma | Skin |
| SKCM-TCGA | Skin_cutaneous_melanoma | Skin |
| STAD-TCGA | Stomach_adenocarcinoma | Stomach |
| TGCT-TCGA | Testicular_germ_cell_tumor | Testis |
| THCA-CN-ICGC | Thyroid_carcinoma | Head_and_neck |
| THCA-SA-ICGC | Thyroid_carcinoma | Head_and_neck |
| THCA-TCGA | Thyroid_carcinoma | Thyroid |
| THYM-TCGA | Thymoma | Thymus |
| UCEC-TCGA | Uterine_corpus_endometrial_carcinoma | Uterus |
| UCS-TCGA | Uterine_carcinosarcoma | Uterus |
| UCS-FR-ICGC | Uterine_carcinosarcoma | Uterus |
| UVM-TCGA | Uveal_melanoma | Skin |
| WT-TARGET | Wilms_Tumor | Kidney |
Citation
To support the continued development of this resource, please cite the following paper if you use DriverDBv4 in your work.
Chia-Hsin Liu , Yo-Liang Lai , Pei-Chun Shen , Hsiu-Cheng Liu , Meng-Hsin Tsai , Yu-De Wang , Wen-Jen Lin , Fang-Hsin Chen , Chia-Yang Li , Shu-Chi Wang , Mien-Chie Hung , Wei-Chung Cheng (2024).
DriverDBv4: a multi-omics integration database for cancer driver gene research. Nucleic Acids Research, 52(D1), D1246–D1252.
DOI Download citation (.nbib)Contact
Copyright© 2010-2025. All Rights Reserved. ©版權所有. Ver. 1.00.003未經允許請勿任意轉載、複製或做商業用途