DriverDBv4: A database for human cancer driver gene research
What is DriverDBv4?
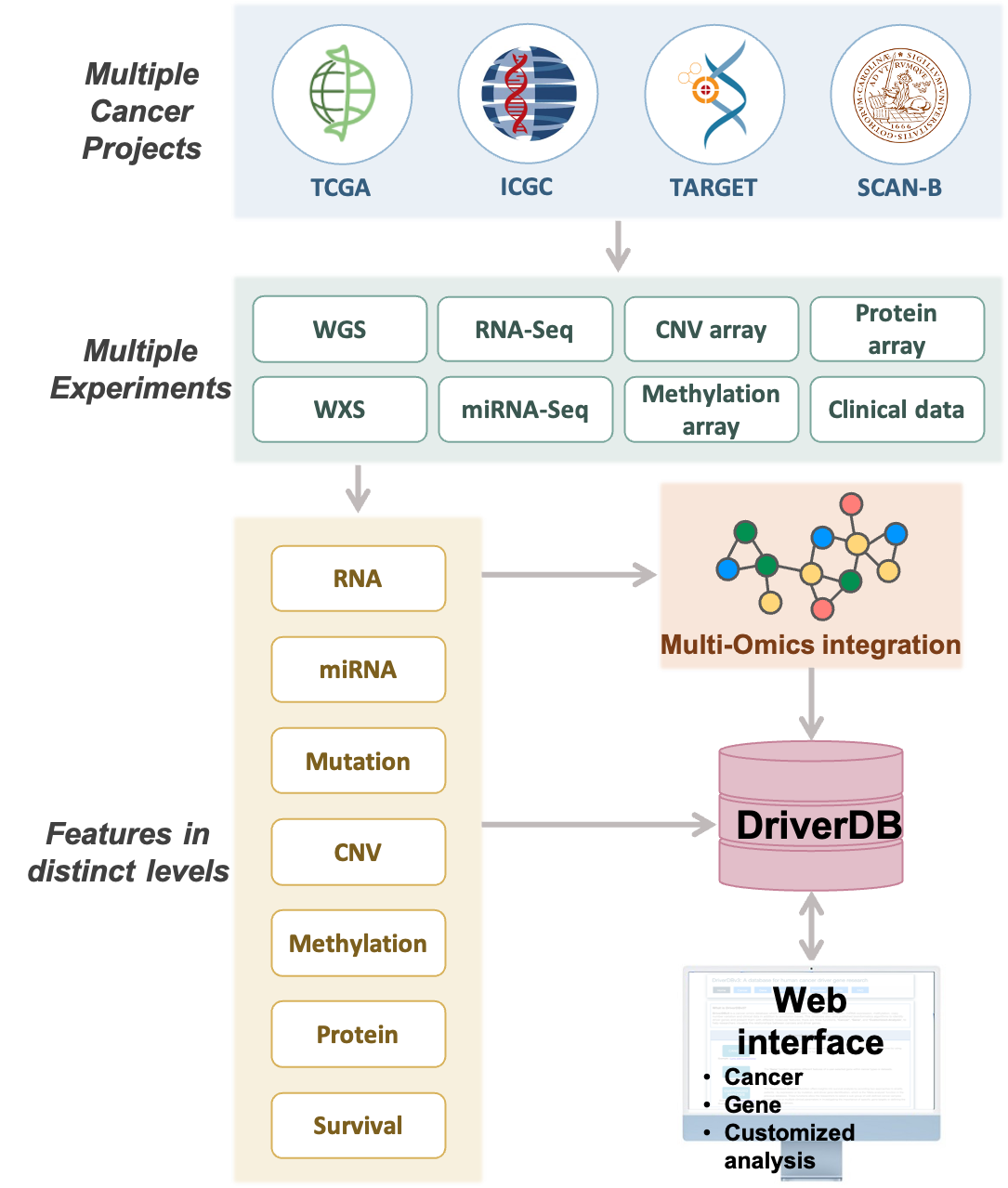
DriverDBv4 is a comprehensive cancer omics database designed to identify and characterize driver genes across mutiple cancer types.
It integrates diverse molecular data including somatic mutations, RNA expression, miRNA expression, protein expression, DNA methylation, and copy number variations alongside clinical information and curated annotations.
Through three intuitive interfaces (Cancer, Gene, and Customized Analysis), researchers can explore complex relationships between specific cancers and their associated driver genes, enabling deeper insights into cancer biology and mechanisms.
Supported browsers: Chrome, Edge, and Firefox.
Search by Functions
The ‘Cancer’ function explores driver genes and miRNA drivers associated with the selected cancer type and their functional roles across multiple omics layers.
The ‘Gene’ function explores a selected gene across cancers, integrating expression, mutation, CNV, methylation, protein, miRNA regulation, survival, and multi-omics evidence to characterize its potential driver roles.
The ‘Customized-Analysis’ function provides five analyses: subgroup analysis, survival analysis, multi-omics driver analysis, signature identification and Multi-variate survival analysis.
- Subgroup Comparison Analyses compare gene-level molecular features, including expression, mutation, CNV, and methylation, across clinically defined patient subgroups to identify biologically or clinically meaningful differences.
- Survival Analyses evaluate how gene expression or mutation status affects survival outcomes in user-defined patient subpopulations using Kaplan–Meier curves and statistical testing.
- Multi-omics Driver Analysis identifies multi-omics driver events that differentiate two patient groups and explore cross-layer regulatory relationships to uncover subgroup-specific drivers.
- Prognostic Signature Identification generates prognostic gene signatures using LASSO and Random Forest, producing performance metrics, ROC curves, heatmaps, and survival plots based on an input gene list.
- Multivariate Survival Analysis constructs multivariable Cox proportional hazards models integrating gene features with clinical variables to assess independent prognostic contributions.
News & Updates
- 2025.08.29 Enhanced high-resolution download feature, available for citation
- 2025.07.29 Launch three new modules in Customized-analysis
- 2023.11.13 Publish DriverDBv4 in Nucleic Acids Res.
- 2023.08.04 Launch DriverDBv4
- 2019.11.08 Publish DriverDBv3 in Nucleic Acids Res.
- 2019.07.22 Launch DriverDBv3
- 2015.12.03 Publish DriverDBv2 in Nucleic Acids Res.
- 2015.08.18 Launch DriverDBv2
- 2013.11.07 Publish DriverDBv1 in Nucleic Acids Res.
- 2013.07.13 Launch DriverDBv1
Citation:
Chia-Hsin Liu, Yo-Liang Lai, Pei-Chun Shen, Hsiu-Cheng Liu, Meng-Hsin Tsai, Yu-De Wang, Wen-Jen Lin, Fang-Hsin Chen, Chia-Yang Li, Shu-Chi Wang, Mien-Chie Hung, Wei-Chung Cheng, DriverDBv4: a multi-omics integration database for cancer driver gene research, Nucleic Acids Research, 2023;, gkad1060, https://doi.org/10.1093/nar/gkad1060
Schematic representation of DriverDBv4
Citation
To support the continued development of this resource, please cite the following paper if you use DriverDBv4 in your work.
Chia-Hsin Liu , Yo-Liang Lai , Pei-Chun Shen , Hsiu-Cheng Liu , Meng-Hsin Tsai , Yu-De Wang , Wen-Jen Lin , Fang-Hsin Chen , Chia-Yang Li , Shu-Chi Wang , Mien-Chie Hung , Wei-Chung Cheng (2024).
DriverDBv4: a multi-omics integration database for cancer driver gene research. Nucleic Acids Research, 52(D1), D1246–D1252.
DOI Download citation (.nbib)Contact
Tel: +886-4-2205-3366 #7929
Email:
wccheng@mail.cmu.edu.tw
Address: 5F., No. 350, Sec. 1, Jingmao Rd., Beitun Dist., Taichung City 406040, Taiwan
BioinfOMICS LAB:
http://www.bioinfomics.org/
Copyright© 2010-2025. All Rights Reserved. ©版權所有. Ver. 1.00.003未經允許請勿任意轉載、複製或做商業用途